Combining de Bruijn graph, overlap graph and microassembly for de novo genome assembly
- 1. Combining de Bruijn graph, overlap graph and microassembly for de novo genome assembly A. Alexandrov, S. Kazakov, S. Melnikov, A. Sergushichev, P. Fedotov, F. Tsarev, A. Shalyto Genome Assembly Algorithms Laboratory St. Petersburg National Research University of Information Technologies, Mechanics and Optics Kazan, 23 Nov 2012
- 2. Algorithm De Bruijn graph Error Quasi- Initial Contig correction contig contig micro- Scaffolding assembly assembly assembly Overlap graph 2
- 3. Error correction • K-mers – substrings of length k. • “Trusted” and “untrusted” k-mers. • Replace “untrusted” k-mers with the “trusted” ones. • If all the k-mers don’t fit into memory. • Divide them into buckets. • Process the buckets independently. 3
- 4. Quasicontig assembly ATGC ??? GTCC ATGC ATGCATGCAGTG GTCC 4
- 5. De Bruijn graph 5
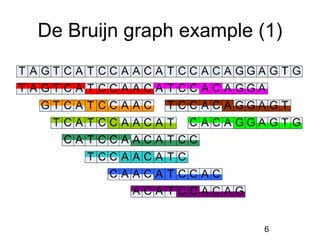
- 6. De Bruijn graph example (1) 6
- 7. De Bruijn graph example (2) GTC TCA CAT ATC TCC AGT GTG CCA CAC CAA GAG GGA AGG CAG ACA AAC 7
- 8. Quasicontig assembly • Build the de Bruijn graph. • For each pair of reads (r1, r2) find the path between the first k-mer of r1 and the last k- mer of r2. • The path has to be of appropriate length. • The path has to be unique. 8
- 9. De Bruijn graph example (3) 9
- 10. De Bruijn graph example (4) 10
- 11. Unique paths correspond to quasicontigs
- 12. Initial contig assembly • Overlap – Suffix array – Inexact overlaps • Layout – Overlap graph • Consensus 12
- 13. Contig microassembly • There are paired reads that map to different contigs. • There are pairs of reads, one of which maps to one of the contigs and the other one maps to the gap between the contigs. 13
- 14. Contig microassembly algorithm • Use Bowtie to find the positions of reads in contigs. • Find all the pairs of contigs connected by many reads. • Build the de Bruijn graph using the reads that map to at least one of the chosen contigs. • Use the quasicontig assembly algorithm to fill the gap. 14
- 15. Results • E. Coli genome – 4,5 million nucleotides. • SRR001665 library, fragment size – 200, read length – 36, coverage – 160. • Before microassembly – 525 contigs, N50 = 17804. • After microassembly – 247 contigs, N50 = 53720. • ABySS – 632 contigs, N50 = 64280. 15
- 17. Acknowledgements • K. Skryabin, E. Prokhorchuk from “Bioengineering” center, for introduction to bioinformatics. • D. Alexeev, from NRI PCM, for the invitation to this conference. 17