15 arrays
- 2. White lines correspond to metal electrodes that connect to individual nanowire devices. The position of the microfluidic channel used to deliver sample is highlighted in blue and has a total size of 6 mm × 500 μm, length × width. The image field is 4.4 × 3.5 mm. C) Scanning electron microscopy image of one (B) Optical image of one row of silicon nanowire device. addressable device elements from the The electrode contacts are region highlighted by the red-dashed visible at the upper right box in A. The red arrow highlights the and lower left regions of the position of a device. The image field is image. (Scale bar: 500 nm.) 500 × 400 μm.
- 3. Bio-chips • Portable, • low cost in high volumes, • low power, • can be integrated with other components Chii-Wann Lin et al, DEVELOPMENT OF MICROMACHINED ELECTROCHEMICAL SENSOR AND PORTABLE METER SYSTEM, a Proceedings of the 20th Annual International Conference of the IEEE 3 Engineering in Medicine and Biology Society, Vol. 20, No 4,1998
- 4. System architectures • Chips – flat platforms, sensors below or above the chip T. Vo-Dinh et al. , Sensors and Actuators, B 74 (2001) 2-11 4
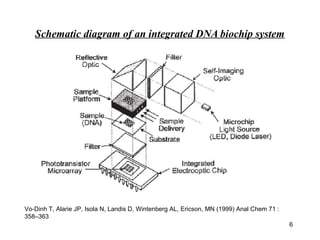
- 6. Schematic diagram of an integrated DNA biochip system Vo-Dinh T, Alarie JP, Isola N, Landis D, Wintenberg AL, Ericson, MN (1999) Anal Chem 71 : 358–363 6
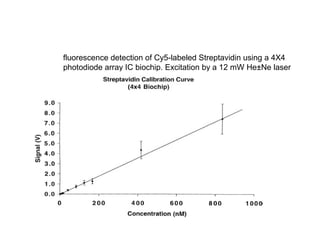
- 7. fluorescence detection of Cy5-labeled Streptavidin using a 4X4 photodiode array IC biochip. Excitation by a 12 mW He±Ne laser (632.8 nm).
- 8. Single detectors vs. Vectors and arrays Single Vector Array 8
- 9. MICROARRAYS It is a 2D array on a solid substrate (usually a glass slide or silicon thin- film cell) that assays large number of biological material using high- throughput screening methods. Types of microarrays include: • DNA microarrays, • oligonucleotide microarrays • MMChips, for surveillance of microRNA populations • Protein microarrays • Tissue microarrays • Cellular microarrays (also called transfection microarrays) • Chemical compound microarrays • Antibody microarrays • Carbohydrate arrays (glycoarrays)
- 10. DNA Arrays (Gene chips)
- 11. Example of a DNA Array (note green, yellow red colors; also note that only part of the total array is depicted)
- 12. 41,000+ unique human genes Example of a DNA Array (note green, yellow red colors; and transcripts represented, all also note that only part of the total with public domain annotations array is depicted) http://www.biomed.miami.edu/arrays/images/agilent_array.jpg
- 13. an arrayed series of thousands of microscopic spots of DNA oligonucleotides, called probes, each containing picomoles of a specific DNA sequence. This can be a short section of a gene or other DNA element that are used as probes to hybridi a cDNA or cRNA sample (called target) the probes are attached to a solid surface by a covalent bond to a chemical matrix (via epoxy-silane, amino-silane, lysine, polyacrylamide or others). The solid surface can be glass or a silicon chip
- 14. • Probe-target hybridization is usually detected and quantified by detection of fluorophore-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. Since an array can contain tens of thousands of probes, a microarray experiment can accomplish many genetic tests in parallel.
- 15. • Colloquially known as an Affy chip when an Affymetrix chip is used. Other microarray platforms, such as Illumina, use microscopic beads, instead of the large solid support.
- 16. • DNA microarrays can be used to measure changes in gene expression levels, to detect single nucleotide polymorphisms (SNPs) , to genotype or resequence mutant genomes.
- 17. Step 1: Create a DNA array (gene “chip”) by placing single-stranded DNA/ Oligonucleotides for each gene to be assayed into a separate “well” on the chip.
- 18. DNA Array: Single-stranded copy DNA Oligonucleotides for each gene in a different well. cDNA cDNA cDNA cDNA cDNA gene 1 gene 2 gene 3 gene 4 gene 5
- 19. Step 2: Extract mRNA from biological tissues subjected to an experimental treatment and from the same tissue subjected to a control treatment. Or from normal and from pathological tissue
- 20. • Step 3- Make single-stranded DNA from the mRNA using “color coded” nucleotides.
- 21. Extract mRNA from Extract mRNA from Control Cells Experimental/pathological Cells Make single-stranded cDNA Make single-stranded cDNA using green nucleotides (e.g. using red nucleotides (e.g. Quantum dots) Quantum dots) cDNA = complementary DNA (DNA synthesized from RNA)
- 22. Step 4: After making many DNA copies of the RNA, extract an equal amount of cDNA from the controls & experimentals and place it into a container.
- 23. Control cDNA Experimental cDNA
- 24. Step 5: Extract a small amount in a pipette.
- 25. Step 6: Insert into first well.
- 26. Step 7: Extract … insert into more cDNA and … second well, etc.
- 27. Step 8: Continue until all wells are filled.
- 28. Step 9: Allow to hybridize, then wash away all single-stranded DNA.
- 29. Result: (1) Some wells have no color-coded cDNA (no mRNA in either type of cell) (2) Some wells have only red (i.e., expressed only in experimental cells) (3) Some wells have only green (i.e., expressed only in control cells) (4) Some wells have both red and green in various mixtures (expressed in both experimental and control cells)
- 30. Step 10: Scan with a laser set to detect the color & process results on computer.
- 31. Results: The colors denote the degree of expression in the experimental versus the control cells. Gene not expressed in control or in experimental cells Only in Mostly in Mostly in Only in Same in control control experimental experimental both cells cells cells cells cells
- 34. Part1 Protein Microarray 1. High throughput analysis of hundreds of thousands of proteins. 2. Proteins are immobilized on glass chip. 3. Various probes (protein, lipids, DNA, peptides, etc) are used.
- 35. Protein Array VS DNA Microarray Target: Proteins DNA (Big, 3D) (Small, 2D) Binding: 3D affinity 2D seq Stability: Low High Surface: Glass Glass Printing: Arrayer Arrayer Amplification: Cloning PCR
- 36. Protein Array Fabrication Protein substrates Polyacrylamide or agarose gels Glass Nanowells Proteins deposited on chip surface by robots Benfey & Protopapas, 2005
- 37. Protein Attachment Diffusion Protein suspended in Diffusion random orientation, but presumably active Adsorption/ Adsorption/Absorption Absorption Some proteins inactive Covalent attachment Covalent Some proteins inactive Affinity Orientation of protein Affinity precisely controlled Benfey & Protopapas, 2005
- 38. Protein Interactions Different capture molecules Antigen– must be used to study antibody different interactions Examples Protein– Antibodies (or antigens) for protein detection Proteins for protein-protein Aptamers interaction Enzyme– Enzyme-substrate for substrate biochemical function Receptor– ligand Benfey & Protopapas, 2005
- 39. Expression Array Probes (antibody) on surface recognize target proteins. Identification of expressed proteins from samples. Typical quantification method for large # of expressed proteins.
- 44. Interaction Array Probes (proteins, peptides, lipids) on surface interact with target proteins. Identification of protein interactions. High throughput discovery of interactions .
- 45. Functional Array Probes (proteins) on surface react with target molecules . Reaction products are detected. Main goal of proteomics.
- 53. Technical Challenges in Protein Chips 1. Poor control of immobilized protein activity. 2. Low yield immobilization. 3. High non-specific adsorption. 4. Fast denaturation of Protein. 5. Limited number of labels – low mutiplexing