Biomedical literature mining
- 1. Biological Literature Mining Lars Juhl Jensen EMBL
- 2. Why?
- 3. Overview Information retrieval and text categorization Methodologies for finding and classifying texts Entity recognition and information extraction Identification of gene/protein/drug names in text Statistical and NLP methods for relation extraction Text- and data-mining Making discoveries from text alone Integration of text and other data types
- 4. Status IR, ER, and simple IE methods are fairly well established Advanced NLP-based IE systems are rapidly being improved Methods for text mining and text/data integration are still in their infancy
- 5. Evaluation Computational linguist lingo Recall = sensitivity Precision = specificity F-score = 2 recall precision/(recall+precision) Best F-score best method CASP-like assessments IR: TREC ER: BioCreAtIvE task 1 (IE: BioCreAtIvE task 2)
- 6. Corpora Plain text Publication abstracts: M EDLINE Full text papers: PubMed Central / Highwire Press Gene summaries: SGD, The Interactive Fly, OMIM, … Patents descriptions: various patent databases Tagged text Categorization: M EDLINE MeSH terms Syntactic tagging: G ENIA Semantic tagging: G ENETAG
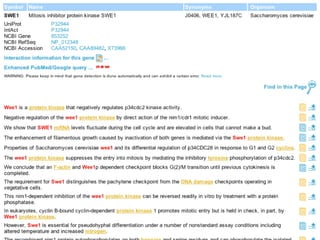
- 7. Example Mitotic cyclin (Clb2)-bound Cdc28 (Cdk1 homolog) directly phosphorylated Swe1 and this modification served as a priming step to promote subsequent Cdc5-dependent Swe1 hyperphosphorylation and degradation
- 8. Information Retrieval and Text Categorization Lars Juhl Jensen EMBL
- 9. Overview Ad hoc information retrieval The user enters a query/a set of keywords The system attempts to retrieve the relevant texts from a large text corpus (typically Medline) Text categorization A training set of texts is created in which texts are manually assigned to classes (often only yes/no) A machine learning methods is trained to classify texts This method can subsequently be used to classify a much larger text corpus
- 10. Ad hoc IR These systems are very useful since the user can provide any query The query is typically Boolean ( yeast AND cell cycle ) A few systems instead allow the relative weight of each search term to be specified by the user The art is to find the relevant papers even if they do not actually match the query Ideally our example sentence should be extracted by the query yeast cell cycle although none of these words are mentioned
- 17. Automatic query expansion In a typical query, the user will not have provided all relevant words and variants thereof By automatically expanding queries with additional search terms, recall can be improved Stemming removes common endings ( yeast / yeasts ) Thesauri can be used to expand queries with synonyms and/or abbreviations ( yeast / S. cerevisiae ) The next logical step is to use ontologies to make complex inferences ( yeast cell cycle / Cdc28 )
- 19. Document similarity The similarity of two documents can be defined based on their word content Each document can be represented by a word vector Words should be weighted based on their frequency and background frequency The most commonly used scheme is tf*idf weighting Document similarity can be used in ad hoc IR Rather than matching the query against each document only, the N most similar documents are also considered
- 20. Document clustering Unsupervised clustering algorithms can be applied to a document similarity matrix All pairwise document similarities are calculated Clusters of “similar documents” can be constructed using one of numerous standard clustering methods Practical uses of document clustering The “related documents” function in PubMed Logical organization of the documents found by IR
- 21. Text categorization These systems are a lot less flexible than ad hoc systems but can attain better accuracy Works on a pre-defined set of document classes Each class is defined by manually assigning a number of documents to it Method Rules may be manually crafted based on a very small set of manually classified documents Statistical machine learning methods can be trained on a large number of classified documents
- 22. Example Mitotic cyclin (Clb2)-bound Cdc28 (Cdk1 homolog) directly phosphorylated Swe1 and this modification served as a priming step to promote subsequent Cdc5-dependent Swe1 hyperphosphorylation and degradation Hints in the text Strong: Cdc28 and Swe1 (“cell cycle” and “yeast”) Weaker: mitotic cyclin , Clb2 , and Cdk1 ( “cell cycle)
- 23. Machine learning Input features Word content or bi-/tri-grams Part-of-speech tags Filtering (stop words, part-of-speech) Singular value decomposition Training Support vector machines are best suited Choice of kernel function Separate training and evaluation sets, cross validation
- 25. Summary Pros and cons of ad hoc IR systems Highly flexible as it is not limited by a training data set Can be very fast if the corpus is properly indexed The accuracy and recall depends strongly on the ability of the user to select the right keywords Some topics are not easily described by a query Pros and cons of text categorization methods Very high accuracy and recall can be attained Requires a separate training set for each category
- 26. Entity Recognition and Information Extraction Lars Juhl Jensen EMBL
- 27. Overview Entity recognition (ER) Finding the genes/proteins/drugs mentioned in a text Word sense disambiguation Information extraction (IE) Simple statistical co-occurrence methods Combining co-occurrence and text categorization Natural Language Processing (NLP)
- 28. Entity recognition An important but boring problem The genes/proteins/drugs mentioned within a given text must be identified Recognition vs. identification Recognition: find the words that are names of entities Identification: figure out which entities they refer to Recognition without identification is of limited use
- 29. Example Mitotic cyclin ( Clb2 )-bound Cdc28 (Cdk1 homolog) directly phosphorylated Swe1 and this modification served as a priming step to promote subsequent Cdc5 -dependent Swe1 hyperphosphorylation and degradation Entities identified S. cerevisiae proteins: Clb2 (YPR119W), Cdc28 (YBR160W), Swe1 (YJL187C), and Cdc5 (YMR001C)
- 30. Recognition Features Morphological: mixes letters and digits or ends on -ase Context: followed by “protein” or “gene” Grammar: should occur as a noun Methodologies Manually crafted rule-based systems Machine learning (SVMs) But what can it be used for?
- 31. Identification A good synonyms list is the key Combine many sources Curate to eliminate stop words Flexible matching to handle orthographic variation Case variation: CDC28 , Cdc28 , and cdc28 Prefixes: myc and c-myc Postfixes: Cdc28 and Cdc28p Spaces and hyphens: cdc28 and cdc-28 Latin vs. Greek letters: TNF-alpha and TNFA
- 32. Disambiguation The same word may mean many different things Entity names may also be common English words ( hairy ) or technical terms ( SDS ) Protein names may refer to related or unrelated proteins in other species ( cdc2 ) The meaning can be resolved from the context ER can distinguish between names and common words Disambiguating non-unique names is a hard problem Ambiguity between orthologs can be safely be ignored
- 38. Co-occurrence Relations are extracted for co-occurring entities Relations are always symmetric The type of relation is not given Scoring the relations More co-occurrences more significant Ubiquitous entities less significant Same sentence vs. same paragraph Simple, good recall, poor precision
- 39. Example Mitotic cyclin ( Clb2 )-bound Cdc28 (Cdk1 homolog) directly phosphorylated Swe1 and this modification served as a priming step to promote subsequent Cdc5 -dependent Swe1 hyperphosphorylation and degradation Relations Correct: Clb2–Cdc28 , Clb2–Swe1 , Cdc28–Swe1 , and Cdc5–Swe1 Wrong: Clb2–Cdc5 and Cdc28–Cdc5
- 41. Categorization Extracting specific types of relations Text categorization methods can be used to identify sentences that mention a certain type of relations Filtering can be done before or after relation extraction Well suited for database curation Text categorization can be reused High recall is most important Curators can compensate for the lack of precision
- 43. NLP Information is extracted based on parsing and interpreting phrases or full sentences Good at extracting specific types of relations Handles directed relations Complex, good precision, poor recall
- 44. Example Mitotic cyclin ( Clb2 )-bound Cdc28 (Cdk1 homolog) directly phosphorylated Swe1 and this modification served as a priming step to promote subsequent Cdc5 -dependent Swe1 hyperphosphorylation and degradation Relations: Complex: Clb2–Cdc28 Phosphorylation: Clb2 Swe1 , Cdc28 Swe1 , and Cdc5 Swe1
- 45. Architecture Tokenization Entity recognition with synonyms list Word boundaries (multi words) Sentence boundaries (abbreviations) Part-of-speech tagging TreeTagger trained on G ENIA Semantic labeling Dictionary of regular expressions Entity and relation chunking Rule-based system implemented in CASS
- 46. Semantic labeling Gene and protein names Cue words for entity recognition Cue words for relation extraction Named entity chunking A CASS grammar recognizes noun chunks related to gene expression: [ nxgene The GAL4 gene ] Relation chunking Our CASS grammar also extracts relations between entities: [ nxexpr T he expression of [ nxgene the cytochrome genes [ nxpg CYC1 and CYC7 ]]] is controlled by [ nxpg HAP1 ]
- 47. [ expression_repression_active Btk regulates the IL-2 gene ] [ dephosphorylation_nominal Dephosphorylation of Syk and Btk mediated by SHP-1 ] [ phosphorylation_nominal phosphorylation of Shc by the hematopoietic cell-specific tyrosine kinase Syk ] [ phosphorylation_nominal the phosphorylation of the adapter protein SHC by the Src-related kinase Lyn ] [ phosphorylation_active Lyn also participates in [ phosphorylation the tyrosine phosphorylation and activation of syk ]] [ phosphorylation_active Lyn , [ negation but not Jak2 ] phosphorylated CrkL ] [ phosphorylation_active Lyn , [ negation but not Jak2 ] phosphorylated CrkL ] [ phosphorylation_active Lyn also participates in [ phosphorylation the tyrosine phosphorylation and activation of syk ]] [ phosphorylation_nominal the phosphorylation of the adapter protein SHC by the Src-related kinase Lyn ] [ phosphorylation_nominal phosphorylation of Shc by the hematopoietic cell-specific tyrosine kinase Syk ] [ dephosphorylation_nominal Dephosphorylation of Syk and Btk mediated by SHP-1 ] [ expression_repression_active IL-10 also decreased [ expression mRNA expression of IL-2 and IL18 cytokine receptors] [ expression_repression_active IL-10 also decreased [ expression mRNA expression of IL-2 and IL18 cytokine receptors ] [ expression_activation_passive [ expression IL-13 expression] induced by IL-2 + IL-18 ] [ expression_activation_passive [ expression IL-13 expression ] induced by IL-2 + IL-18 ] [ expression_repression_active Btk regulates the IL-2 gene ]
- 49. MedScan
- 50. Summary Entity recognition The best methods rely on curated synonyms lists Co-occurrence methods High recall but typically poor accuracy Cannot deal with directed interactions Natural language processing High accuracy but typically poor recall Rule development is time consuming
- 51. Text- and Data-mining Lars Juhl Jensen EMBL
- 52. Overview Pure text-mining Discovery of global trends Inference of overlooked relations Integration of text and other data sources Augmented text-mining methods Automated annotation of high-throughput data
- 53. Trends Most similar to existing data mining approaches Although all the detailed data is in the text, people may have missed the big picture Temporal trends Historical summaries Forecasting Correlations “ Customers who bought this item also bought …”
- 54. Time
- 55. Successful genes
- 56. Buzzwords
- 57. Correlations “ Customers who bought this item also bought …” Protein networks “ Proteins that regulate expression …” “ Proteins that control phosphorylation …” “ Proteins that are phosphorylated …” Co-author networks
- 58. Transcriptional networks 32 79 83 3592 Regulates Regulated P < 9 10 -9
- 59. Signaling pathways 11 27 44 3704 Phosphorylates Phosphorylated P < 2 10 -7
- 60. Multiple regulation 8 107 47 3625 Expression Phosphorylation P < 5 10 -4
- 62. Nuggets New relations can be inferred from published ones This can lead to actual discoveries if no person knows all the facts required for making the inference Combining facts from disconnected literatures Swanson’s pioneering work Fish oil and Reynaud's disease Magnesium and migraine
- 65. Integration Automatic annotation of high-throughput data Loads of fairly trivial methods Protein interaction networks Can unify many types of interactions Powerful as exploratory visualization tools More creative strategies Identification of candidate genes for genetic diseases Linking genes to traits based on species distributions
- 71. RCCs
- 72. Disease candidate genes Rank the genes within a chromosomal region to which a disease has been mapped Methods G2D Gene Function Chemical Phenotype Disease Uses M EDLINE but not the text B ITOLA Gene Words Disease (similar to A RROWSMITH ) Hide and co-workers Gene Tissue Disease
- 73. G2D
- 77. Genotype–phenotype Genes can be linked to traits by comparing the species distributions of both Mainly works for prokaryotes Traits are represented by keywords Finding the species profiles Gene profiles are found by sequence similarity Keyword profiles are based co-occurrence with the species name in M EDLINE
- 80. Annotation High-throughput experiments of result in groups of related genes ER is used to find the associated abstracts The frequency of each word is counted in the abstracts Background frequencies of all words are pre-calculated A statistical test is used to rank the words (typically Fisher’s exact test) The same strategy can be applied to find MeSH terms associated with a gene cluster
- 81. Summary Mining for overlooked relations Few overlooked relations can be found from text alone Methods that combine text and other data types have much better discovery potential Annotation/interpretation of high-throughput data Molecular networks can be useful for gaining an overview of large expression data sets Literature can be used to find keywords for a group of genes, but this has few advantages over using GO terms
- 82. Outlook Lars Juhl Jensen EMBL
- 83. Death? Literature mining will not be made obsolete by <insert your favorite new technology here> Repositories are always made too late There will always be new types of relations Semantically tagged XML may replace ER (hopefully!) Semantically tagged XML will never tag everything Specific IE problems will become obsolete Protein function Physical protein interactions
- 84. Permission denied Open access Literature mining methods cannot retrieve, extract, or correlate information from text unless it is accessible Restricted access is already now the primary problem Standard formats Getting the text out of a PDF file is not trivial Many journals now store papers in XML format Where do I get all the patent text?!
- 85. Innovation The basic tools are now in place for IR, ER, and IE Development was driven by computational linguists Text- and data-mining Biologists are needed Collaboration with linguists Lack of innovation Very few new ideas Text should be combined with other data
- 86. Acknowledgments EML Research Jasmin Saric Isabel Rojas EMBL Heidelberg Peer Bork Miguel Andrade Rossitza Ouzounova Jan Korbel Tobias Doerks
- 87. Exercises Lars Juhl Jensen EMBL
- 88. Information retrieval PubFinder http://www.glycosciences.de/tools/PubFinder/ Ideas Do a very specific search on PubMed that retrieves only around 10–20 relevant papers See if PubFinder is able to retrieve more Compare this with using the “Related Articles” function in PubMed
- 89. Entity recognition iHOP http://www.pdg.cnb.uam.es/UniPub/iHOP/ Ideas Compare iHOP vs. PubMed for finding papers related to a particular gene Use iHOP to construct a small literature-based network
- 90. Information extraction Relation extraction iProLINK ( http://pir.georgetown.edu/iprolink/ ) PreBIND ( http://prebind.bind.ca ) PubGene ( http://www.pubgene.org ) Ideas Check how complex sentences iProLINK can handle Check how well PreBIND can discriminate between physcial and other interactions (other interactions can be found with PubGene, ProLinks, or STRING)
- 91. Text mining A RROWSMITH http://arrowsmith.psych.uic.edu Ideas Fish oil and Reynaud's disease Magnesium and migraine Arginine and somatomedin C Estrogen and Alzheimer's disease
- 92. Integration 1 Protein networks S TRING beta version ( http://string.embl.de :8080 ) ProLinks ( http://dip.doe-mbi.ucla.edu/pronav/ ) Ideas Use both tools to find functions for proteins of known and unknown function Use S TRING to construct a network for a set of proteins Try to reproduce the Ssn3–Msn2–Hsp104 link
- 93. Integration 2 Finding candidate disease genes G2D ( http://www.ogic.ca/projects/g2d_2/ ) B ITOLA ( http://www.mf.uni-lj.si/bitola/ ) Ideas Take a look at the G2D results for some diseases where you know which types of genes would be sensible to suggest Compare the results with B ITOLA (if you have the patience to figure out there interface!)
- 94. Integration 3 Annotation of expression data MedMiner ( http://discover.nci.nih.gov/textmining/ ) Ideas Stating the obvious … do the one thing that MedMiner can do …













































![Semantic labeling Gene and protein names Cue words for entity recognition Cue words for relation extraction Named entity chunking A CASS grammar recognizes noun chunks related to gene expression: [ nxgene The GAL4 gene ] Relation chunking Our CASS grammar also extracts relations between entities: [ nxexpr T he expression of [ nxgene the cytochrome genes [ nxpg CYC1 and CYC7 ]]] is controlled by [ nxpg HAP1 ]](https://image.slidesharecdn.com/jen05talk8-1217968131114198-8/85/Biomedical-literature-mining-46-320.jpg)
![[ expression_repression_active Btk regulates the IL-2 gene ] [ dephosphorylation_nominal Dephosphorylation of Syk and Btk mediated by SHP-1 ] [ phosphorylation_nominal phosphorylation of Shc by the hematopoietic cell-specific tyrosine kinase Syk ] [ phosphorylation_nominal the phosphorylation of the adapter protein SHC by the Src-related kinase Lyn ] [ phosphorylation_active Lyn also participates in [ phosphorylation the tyrosine phosphorylation and activation of syk ]] [ phosphorylation_active Lyn , [ negation but not Jak2 ] phosphorylated CrkL ] [ phosphorylation_active Lyn , [ negation but not Jak2 ] phosphorylated CrkL ] [ phosphorylation_active Lyn also participates in [ phosphorylation the tyrosine phosphorylation and activation of syk ]] [ phosphorylation_nominal the phosphorylation of the adapter protein SHC by the Src-related kinase Lyn ] [ phosphorylation_nominal phosphorylation of Shc by the hematopoietic cell-specific tyrosine kinase Syk ] [ dephosphorylation_nominal Dephosphorylation of Syk and Btk mediated by SHP-1 ] [ expression_repression_active IL-10 also decreased [ expression mRNA expression of IL-2 and IL18 cytokine receptors] [ expression_repression_active IL-10 also decreased [ expression mRNA expression of IL-2 and IL18 cytokine receptors ] [ expression_activation_passive [ expression IL-13 expression] induced by IL-2 + IL-18 ] [ expression_activation_passive [ expression IL-13 expression ] induced by IL-2 + IL-18 ] [ expression_repression_active Btk regulates the IL-2 gene ]](https://image.slidesharecdn.com/jen05talk8-1217968131114198-8/85/Biomedical-literature-mining-47-320.jpg)














































