Lecture 2 Dl.ppt
- 1. Lecture 2 Membrane Structure & Ligand-gated channels In Memory of my good friend and colleague Massimo Sassaroli
- 2. Membrane Structure Learning Objectives Know the major classes of natural lipids, their structural characteristics and the properties of head groups and hydrocarbon chains Understand the thermodynamic basis of lipid and detergent assembly: the hydrophobic effect Understand the physical connection between the shape of lipid molecules and that of the aggregates they form: critical packing parameter Understand the connection between the configuration of acyl chains, their packing and the properties of the bilayer Know the connection between bilayer structure and dynamics and translational diffusion of lipids and proteins. Experimental approach to the measurement of diffusion of membrane components: Fluorescence Recovery After Photobleaching Understand the characteristics of the fluid phase bilayer as shown by diffraction experiments and computer simulations of lipid dynamics
- 3. Cell membranes are dynamic fluid structures. Most of their molecules are able to move about in the plane of the membrane ~30% of the proteins encoded in an animal cell’s genome are membrane proteins
- 5. Acyl Chains
- 6. Head Groups
- 7. Major classes of lipids
- 8. Glycolipids
- 12. A 2 : cleave at C2 e.g. AA to PG and LT A 1 : cleave at C1 C: cleave at C3 e.g. PIP 2 to DAG and IP 3 D: cleave at C3 e.g. phosphatidic acid
- 14. (C)
- 15. G = H T S Hydrophobicity and thermodynamics of lipid assembly van der Waals force electrostatic force hydrophobic force Electrostatic interactions are lost when H 2 O binds to the hydrocarbon chain. This translates to both an enthalpic and entropic (immobilization of H 2 O) energy cost. Tight packing of hydrocarbon chains and release of H 2 O back to the balk reduces G => Hydrophobic effect
- 20. Saturated chains in the all- trans configuration, 19 Å 2 PE S 39 Å 2 PC S 50 Å 2
- 24. Membrane Proteins and Lipid-Protein Interactions Learning Objectives Modes of protein-membrane interaction. Prediction of membrane protein structure and topology: Hydropathy analysis and the ‘positive‑inside’ rule. Lipid‑modifications of proteins: hydrophobicity and membrane-binding affinity Modulation of reversible protein-membrane binding: the myristoyl switch. Phosphoinositide control of Ion Channel Activity
- 44. Monitoring PIP 2 hydrolysis – translocation of PH-GFP Keselman and Logothetis, (2007) Channels 1:113-123
- 45. AASt PI(4,5)P 2 Kir channels run down upon channel excision and are reactivated by PIP 2
- 46. Modulation of Kir2 activity by PLC stimulation diC 8 PI(4,5)P 2
- 47. Channel PIP 2 -sensitive sites are congregated near helix bundle crossing and H-I loop gates
- 48. PIP 2 tethers the N- and C-termini to the inner leaflet of the plasma membrane gating the channel to the open state WORKING HYPOTHESIS
- 49. Kir3 modulation sites congregate around PIP 2 sites Cyan: PIP 2 sites Red: G sites Pink: Na + sites Orange: Phosphorylation sites
- 50. Know the mechanism by which the metabolic state of the cell is coupled to membrane electrical events, such as those leading to secretion of insulin. Know the mechanism of activation of G protein-gated K channels, as an example of a membrane-delimited pathway of regulating the activity of intracellular ligand-gated ion channels. Modulation of Ion channels by soluble second messengers Sensory transduction: Know the role of CNG channels in phototransduction. Understand how the balance of CNG and K channels gives rise to the "dark current", which is inhibited during a light flash. Know the subunit composition of nicotinic ACh channels and general topology of the subunits. Know the general activation mechanism for NMDA and non-NMDA channels and role in LTP. The Transient Receptor Potential (TRP) Channel Family Intracellular and Extracellular Ligand-Gated Ion Channels Learning Objectives
- 51. ATP-Sensitive K + Channels
- 52. Electrostatic Channel-PIP 2 Interactions
- 53. G Protein-sensitive K + channels
- 54. Cytosolic domains of GIRK1 Nishida et al., 2002 Cell 111:957-65
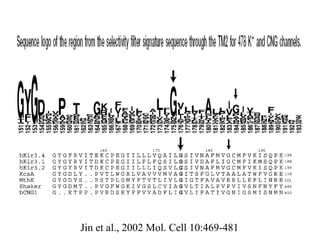
- 55. Jin et al., 2002 Mol. Cell 10:469-481
- 56. G175 mutants of Kir3.4* Jin et al., Mol. Cell 10 , 469 (2002)
- 57. The cells of the Retina
- 58. The “dark” current and the response to light of a photoreceptor cell
- 60. Image reconstruction from cryo-EM
- 61. Two types of Glutamate Receptors
- 63. Glutamate Receptor ligand-binding domain