Lecture 9 slides: Machine learning for Protein Structure ...
- 1. Introduction to Bioinformatics 9. Machine Learning for Protein Structure Prediction #1 Course 341 Department of Computing Imperial College, London © Simon Colton
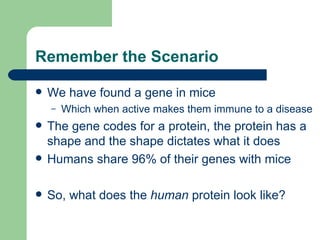
- 2. Remember the Scenario We have found a gene in mice Which when active makes them immune to a disease The gene codes for a protein, the protein has a shape and the shape dictates what it does Humans share 96% of their genes with mice So, what does the human protein look like?
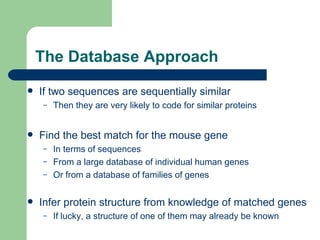
- 3. The Database Approach If two sequences are sequentially similar Then they are very likely to code for similar proteins Find the best match for the mouse gene In terms of sequences From a large database of individual human genes Or from a database of families of genes Infer protein structure from knowledge of matched genes If lucky, a structure of one of them may already be known
- 4. There is another way… Machine learning: general set of techniques For teaching a computer to make predictions By observing given correct predictions (being trained) Special type of prediction Classification of objects into classes E.g., images into faces/cars/landscapes E.g., drugs into toxic/non-toxic ( binary classification) We want to predict a protein’s structure Given its sequence
- 5. A Good Approach Look at regions of a protein i.e., lengths of residues Define ways to describe the regions So that we can infer the structure of a protein From a description of all its regions Learn methods for predicting: What type of region a particular residue will be in Apply this to a protein sequence To find contiguous regions with same description Put regions together to predict entire structure
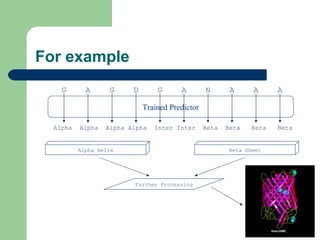
- 6. For example G A G D G A N A A A Alpha Alpha Alpha Alpha Inter Inter Beta Beta Beta Beta Trained Predictor Alpha Helix Beta Sheet Further Processing
- 7. Two Main Questions How do we describe protein structures? What are alpha helices and beta-sheets? Covered in the next lecture How do we train our predictors? Covered in this lecture (and the start of the next…)
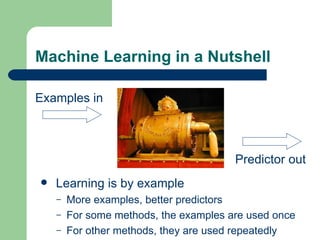
- 8. Machine Learning in a Nutshell Examples in Predictor out Learning is by example More examples, better predictors For some methods, the examples are used once For other methods, they are used repeatedly
- 9. Machine Learning Considerations What is the problem for the predictor to address? What is the nature of our data? How will we represent the predictor? How will we train the predictor? How will we test how good the predictor is?
- 10. Types of Learning Problems in Bioinformatics Class membership e.g., predictive toxicology Prediction of sequences e.g., sequences of protein sub-structures Classification hierarchies e.g., folds, families, super-families Shape descriptions e.g., binding site descriptions Temporal models e.g., activity of cells, metabolic pathways
- 11. Learning Data Data comes in many forms, in particular: Objects (to be classified/predicted for) Classifications/predictions of objects Features of objects (to use in the prediction) Problems with data Imprecise information (e.g., badly recorded data) Irrelevant information (e.g., additional features) Incorrect information (e.g., wrong classifications) Missing classifications Missing features for sets of objects
- 12. Types of Representations Logical Decision trees , grammars, logic programs Symbolic, understandable representations Probabilistic Neural networks , Hidden Markov Models , SVMs Mathematical functions, not easy to understand Mixed Bayesian Networks, Stochastic Logic Programs Have advantages of both, more difficult to work with
- 13. Advantages of Representations Probabilistic models: Can handle noisy and imprecise data Useful when there is a notion of uncertainty (in data/hypothesis) Well-founded (300 years of development) Good statistical algorithms for estimation Logical models Richness of description Extensibility - probabilities, sequences, space, time, actions Clarity of results Well-founded (2400 years of development)
- 14. Decision Tree Representations Input is a set of features Describing an example/situation Many “if-then” choices Leaves are decision Logical representation: “ If then” is implication Branches are conjunctions Different branches comprise A disjunction
- 15. Artificial Neural Networks Layers of nodes Input is transformed into numbers Weighted averages are fed into nodes High or low numbers come out of nodes A Threshold function determines whether high or low Output nodes will “fire” or not Determines classification For an example
- 16. Logic Program Representations Logic programs are a subset of first order logic They consist of sets of Horn clauses Horn clause: A conjunction of literals implying a single literal Can easily be interpreted At the heart of the Prolog programming language
- 17. Learning Decision Trees Problem: what feature do nodes in the tree test? And what happens for each case ID3 algorithm: Uses a notion of “Information gain” Based on entropy: how (dis)organised data is Chooses the node with the highest information gain As the node to add to the tree next Then restricts examples for next node
- 18. Learning Artificial Neural Networks First problem: layer structure Usually done through trial and error Main problem: choosing the weights Uses a back-propagation algorithm to train them Each example is given If currently correctly classified, that’s fine If not, the errors from the output are passed back Propagated in order to change the weights throughout Only very small changes are made (avoid un-doing good work) Once all examples have been given We start again, until some termination conditions (accuracy) met Often requires thousands of such training ‘epochs’
- 19. Learning Logic Programs A notion of generality is used One logic program is more general than another If one follows from another (sort of) [subsumption] A search space of logic programs is defined Constrained using a language bias Some programs search from general to specific Using rules of deduction to go between sentences Other programs search from specific to general Using inverted rules of deduction Search is guided by: Performance of the LP with respect to classifying training examples Information theoretic calculations to avoid over-specialisations
- 20. Testing Learned Predictors #1 Imperative to test on unseen examples Cannot report accuracy on examples which have been used to train the predictor, because the results will be heavily biased Simple method: Hold back When number of examples > 200 (roughly) Split into a training set and a test set (e.g., 80%/20%) Never let the training algorithm see the test set Report the accuracy of the predictor on the test set only Have to worry about statistics with smaller numbers This kind of testing came a little late to bioinformatics Beware conclusions drawn about badly tested predictors
- 21. N-Fold Cross Validation Leave one out For m < 20 examples Train on m-1 examples, test predictor on left out example Do this for every example and report the average accuracy N-fold cross validation Randomly split into n mutually exclusive sets (partitions) For every set S Train using all examples from the other n-1 sets Test predictor on S, record the accuracy Report the average accuracy over all the sets 10-fold cross validation is common
- 22. Testing Learned Predictors #2 Often different consideration for different contexts E.g., false positives/negatives in medical diagnosis Confusion matrix For binary prediction tasks Predicted F Predicted T number = a number = b (false pos) number = c (false neg) number = d Actually F Actually T Let t = a+b+c+d Predictive accuracy = (a+d)/t Majority class = max ((a+b)/t, (c+d)/t) Precision = Selectivity = d/(b+d) Recall = Sensitivity = d/(c+d)
- 23. Comparing Learning Methods A very simple method: Majority class predictor Predict everything to be in the majority class Trained predictors must beat this to be credible N-fold cross validation results are compared To show an advance in predictor technology However, accuracy is not the only consideration Speed, memory and comprehensibility
- 24. Overfitting It’s easy to over-train predictors If a predictor is substantially better for the training set than the test set, it is overfitting Essentially, it has memorised aspects of the examples, rather than generalising properties of them This is bad: think of a completely new example Individual learning schemes have coping methods Easy general approach to avoiding overfitting: Maintain a validation set to perform tests on during training When performance on the validation set degrades, stop learning Be careful of blips in predictive accuracy (leave a while, then come back) Note: this shouldn’t be used as the testing set














![Learning Logic Programs A notion of generality is used One logic program is more general than another If one follows from another (sort of) [subsumption] A search space of logic programs is defined Constrained using a language bias Some programs search from general to specific Using rules of deduction to go between sentences Other programs search from specific to general Using inverted rules of deduction Search is guided by: Performance of the LP with respect to classifying training examples Information theoretic calculations to avoid over-specialisations](https://image.slidesharecdn.com/lecture-9-slides-machine-learning-for-protein-structure3876/85/Lecture-9-slides-Machine-learning-for-Protein-Structure-19-320.jpg)




