Bio-optical Sensing Dissertation
- 1. UT Biomedical Informatics Lab Depth Resolved Diffuse Reflectance Spectroscopy Ricky Hennessy The Biomedical Informatics Lab (BMIL) The Biophotonics Laboratory
- 2. UT Biomedical Informatics Lab Committee 1/60 Mia K. Markey, Ph.D. - Advisor The Biomedical Informatics Lab James W. Tunnell, Ph.D. - Advisor The Biophotonics Lab Stanislav Emelianov, Ph.D. Ultrasound Imaging and Therapeutics Andrew K. Dunn, Ph.D. Functional Optical Imaging Lab Ammar M. Ahmed, M.D. Dermatology at Seton
- 3. UT Biomedical Informatics Lab Diffuse Reflectance Spectroscopy (DRS) 2/60
- 4. UT Biomedical Informatics Lab Applications of DRS 3 Cancer Detection Soil Characterization Wearable Tech Food Quality Endoscopic Surgery Cosmetic Applications
- 5. UT Biomedical Informatics Lab Cancer Detection with DRS 4/60 Extract Features from Data Use Features to Create Classifier Rajaram et al. Lasers Surg Med 42:876-887 (2010)
- 6. UT Biomedical Informatics Lab DRS Instrumentation 5 Spectrometer LightSource SourceFiber DetectorFiber Computer FiberBundle
- 7. UT Biomedical Informatics Lab Light Absorption in Tissue 6/60 400 450 500 550 600 0 0.2 0.4 0.6 0.8 1 WAVELENGTH (nm) ABSORPTION(a.u.) Melanin HbO 2 Hb Other Absorbers • Water • Bilirubin • Lipid • Protein • Collagen
- 8. UT Biomedical Informatics Lab Light Scattering in Tissue 7/60
- 9. UT Biomedical Informatics Lab Biological Origins of Scattering 8/60 Scattering is caused by index of refraction mismatches • Cells = ~10 μm • Nuclei = ~1 μm • Collagen = 0.1 μm • Membranes = 0.01 μm
- 10. UT Biomedical Informatics Lab Scattering Coefficient (μs) 9/60 Scattering coefficient (μs) is proportional to concentration of scatterers in a medium μs -1 is the average distance a photon travels between scattering events μs
- 11. UT Biomedical Informatics Lab Direction of Scattering 10/60 Isotropic Scattering Anisotropic Scattering Tissue scattering is in forward direction (g = ~0.9) Henyey-Greenstein Phase Function g = 0
- 12. UT Biomedical Informatics Lab Radiative Transport Equation (RTE) 11/60 ΔEnergy Scattering in Scattering out Absorption
- 13. UT Biomedical Informatics Lab Reduced Scattering Coefficient 12/60 1 2 10 9 8 7 6 5 43 Using reduced scattering with isotropic scattering is equivalent to larger scattering with anisotropic scattering
- 14. UT Biomedical Informatics Lab Diffusion Approximation to the RTE 13/60 Kienle et al., JOSA A, 1997, 14(1), 246-254 Assumes isotropic scattering • Scattering >> absorption • Source Detector Separation > ~ 1 mm Blood is highly absorbing Epidermis is ~100 μm thick
- 15. UT Biomedical Informatics Lab Monte Carlo Simulation 14/60
- 16. UT Biomedical Informatics Lab 15
- 17. UT Biomedical Informatics Lab 16
- 18. UT Biomedical Informatics Lab The Monte Carlo Lookup Table (MCLUT) Method Hennessy et al., JBO, 2013, 18(3), 037003
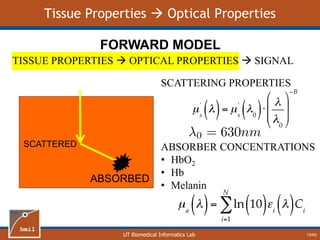
- 19. UT Biomedical Informatics Lab Tissue Properties Optical Properties SCATTERED ABSORBED TISSUE PROPERTIES OPTICAL PROPERTIES SIGNAL FORWARD MODEL SCATTERING PROPERTIES ABSORBER CONCENTRATIONS • HbO2 • Hb • Melanin 18/60
- 20. UT Biomedical Informatics Lab Forward Model Flowchart TISSUE PROPERTIES OPTICAL PROPERTIES SIGNAL FORWARD MODEL Light Transport Model Scattering at λ0, Concentration of chromophores Calculate absorption and scattering coefficients at each wavelength Scattering Absorption Use light transport model to calculate diffuse reflectance Generate diffuse reflectance spectrum 19/60
- 21. UT Biomedical Informatics Lab Monte Carlo on GPU Modern processor w/ 4 cores = 4 times speedup Modern GPU w/ 500 cores = 500 times speedup! < $300 Alerstam et al., Biomed. Opt. Express., 2010, 1, 658-675 20/60
- 22. UT Biomedical Informatics Lab Monte Carlo Lookup Table (1 Layer) 400 450 500 550 600 650 700 0 1 2 3 4 5 6 7 8 9 WAVELENGTH (nm) REFLECTANCE 21/60
- 23. UT Biomedical Informatics Lab MCLUT Inverse Model REFLECTANCE OPTICAL PROPERTIES TISSUE PROPERTIES Optimization Routine 22/60
- 24. UT Biomedical Informatics Lab Calibration MCLUT Modeled Spectra RMC = photons counted Measured Spectra Rmeas = Iraw/Istandard Modeled spectra and measured spectra have same optical properties 23/60
- 25. UT Biomedical Informatics Lab Validation of One-Layer Model RMSPE = 2.42% RMSPE = 1.74% Validation with 3 X 6 matrix of phantoms containing hemoglobin and polystyrene beads. Decreased percent error of 3.16% and 10.86% for μs' and μa, respectively, when compared to experimental LUT method 24/60
- 26. UT Biomedical Informatics Lab Errors Caused by One-Layer Assumption for Skin Hennessy et al., JBO, 2015, 20(2), 027001
- 27. UT Biomedical Informatics Lab Fit Two-Layer Data with One Layer Model 26/60 Hb + HbO2 melanin melanin + Hb + HbO2 1. Create two-layer spectra 2. Fit with one- layer model • [mel] • [Hb] • SO2 • Scattering • Epidermal thickness • [mel] • [Hb] • SO2 • Scattering • Vessel radius Notice that the fit is very good
- 28. UT Biomedical Informatics Lab Melanin 27/60 • One-Layer model underestimates [mel] • Magnitude of error is dependent on epidermal thickness (Z0) • Z0 Error
- 29. UT Biomedical Informatics Lab Hemoglobin 28/60 • One-Layer model underestimates [Hb] • Magnitude of error is dependent on epidermal thickness (Z0) • Z0 Error
- 30. UT Biomedical Informatics Lab Oxygen Saturation 29/60 • One-Layer model overestimates SO2 when SO2 < 50% • One-Layer model underestimates SO2 when SO2 > 50% • Magnitude of error is dependent on epidermal thickness (Z0) • Z0 Error
- 31. UT Biomedical Informatics Lab Pigment Packaging 30/60 • In tissue, blood is confined to vessels • This significantly reduces the optical path length where absorption is high (Soret Band) • Causes a flattening of the absorption spectrum
- 32. UT Biomedical Informatics Lab Vessel Radius vs. Epidermal Thickness 31/60 0 50 100 150 200 250 300 0 50 100 150 200 250 300 350 400 450 500 Top Layer Thickness (mm) VesselRadius(mm) • Vessel radius (pigment packaging) factor is highly correlated with top-layer thickness • Pigment packaging factor is likely a combination of vessel packaging and epidermal thickness
- 33. UT Biomedical Informatics Lab Correlation Between [Hb] and [mel] 32/60 R = 0.04 R = 0.80 One-layer assumption causes artificial correlation between [Hb] and [mel]
- 34. UT Biomedical Informatics Lab Conclusions about One-Layer Errors Causes underestimation of [Hb] and [mel] – Magnitude of error is function of epidermal thickness Causes error in SO2 that is a function of epidermal thickness as well as SO2 Vessel Packaging factor and epidermal thickness are highly correlated Causes an artificial correlation in [Hb] and [mel] 33/60
- 35. UT Biomedical Informatics Lab The Two-Layer Monte Carlo Lookup Table Method Sharma, Hennessy et al., Biomed Optics Express, 2014, 5(1), 40-53
- 36. UT Biomedical Informatics Lab Motivation of Two-Layer Model Epidermis Dermis Stratum Corneum Melanin Hemoglobin 35/60
- 37. UT Biomedical Informatics Lab Motivation of Two-Layer Model • Pigmentary disorder studies • Disease (rosacea, lupus, scleroderma, morphea, lymphederma) monitoring • Treatment outcome measures for many cosmetic procedures • Topical medical absorption studies • Measuring thickness of psoriatic plaque • Determination of epidermal thickness Melasma Method of melasma treatment depends on depth of melanin Wood’s lamp is current method to determine location of melanin Qualitative – More contrast for epidermal melasma. Doesn’t work for patients with dark skin. Asawansa et al., Int. J. Derm, 1999, 38, 801-807 36/60
- 38. UT Biomedical Informatics Lab Two Layer MCLUT We can create a 5D LUT 1. Top layer absorption 2. Bottom layer absorption 3. Scattering 4. Top layer thickness 5. SDS 37/60 Segment of 5D lookup table • Z0 = 200 μm • SDS = 200 μm • R1 = R2 = 100 μm Sharma, Hennessy et al., Biomed. Opt. Express, 2014, 5(1), 40-53
- 39. UT Biomedical Informatics Lab Two-Layer Forward Model 38/60
- 40. UT Biomedical Informatics Lab Two-Layer MCLUT Inverse Model 39/60
- 41. UT Biomedical Informatics Lab Two-Layer Phantom Construction 40/60
- 42. UT Biomedical Informatics Lab Phantom Validation Study PHANTOM μs’ (mm-1) μa,t (mm-1) μa,b (mm-1) 1 1.5 0.25 1.275 2 1.5 0.25 1.275 3 1.5 2.3 0.25 4 1.5 2.3 0.25 5 1.5 0.25 2.3 6 1.5 0.25 2.3 7 2.85 0.25 2.3 8 2.85 0.25 2.3 9 2.85 2.3 0.25 10 2.85 1.275 0.25 11 2.85 1.275 2.3 12 0.75 0.25 1.275 13 0.75 1.275 0.25 14 0.75 0.25 2.3 Set of Two Layer Phantoms Range of scattering and absorption in skin μs’ = 1-3 mm-1 μa = 0 – 2.3 mm-1 Nichols et al., JBO, 2012, 17(5), 057001 41/60
- 43. UT Biomedical Informatics Lab Phantom Data 42/60
- 44. UT Biomedical Informatics Lab Results: Top Layer Thickness 43/60 Error = 10% for Z0 < 500 μm Photons are not sampling this deep.
- 45. UT Biomedical Informatics Lab Results: All Optical Properties 44/60
- 46. UT Biomedical Informatics Lab Results: Dependence on SDS 45/60 Main Takeaway Accuracy of extracted parameters is dependent on probe geometry. This is due to sampling depth of probe.
- 47. UT Biomedical Informatics Lab Sampling Depth of Diffuse Reflectance Spectroscopy Probes Hennessy et al., JBO, 2014, 19(10), 107002
- 48. UT Biomedical Informatics Lab Probe Geometry and Sampling Depth 47 Tissue Source Fiber Detection Fiber SDS Sampling Depth Z(μa,μs’,rS,rD,SDS) rS rD -Tissue Optical Properties -Source/Detector Fiber Sizes -Source/Detector Separation * Credit to Will Goth for this slide 47/60
- 49. UT Biomedical Informatics Lab Defining Sampling Depth 48/60 This experiment was performed computationally (MC simulation) and experimentally (phantoms)
- 50. UT Biomedical Informatics Lab Experimental Validation 49/60 E = 1.71% E = 1.27% E = 1.24% SDS = 370 μm SDS = 740 μm SDS = 1110 μm Look at axes to see deeper sampling for larger SDSs
- 51. UT Biomedical Informatics Lab Analytical Model of Sampling Depth 50/60 This expression can be used to aid in the design of application specific DRS probesE = 2.89% Expression was found using TableCurve 3D. Free parameters [a1, a2, a3, a4] were selected using a least-squares fitting algorithm.
- 52. UT Biomedical Informatics Lab Choice of g and Phase Function 51/60 The choice for g and phase function had negligible impact on the sampling depth model
- 53. UT Biomedical Informatics Lab Applying the Sampling Depth Model 52/60 6-around-1 adjacent fiber orientation with medium (series 1), high (series 2), and low (series 3) absorption. Sampling depth changes with wavelength
- 54. UT Biomedical Informatics Lab Pilot Study: Two-Layer MCLUT Method Applied to In Vivo Data
- 55. UT Biomedical Informatics Lab Study Population and Data 54/60 • 80 Subjects • IRB approval from UT Austin - #00002030 • 51 males, 29 Females • Average age of 25.7 years • Ages 18-46 • Measured spectra from the following anatomical locations 1. Back 2. Calf 3. Cheek 4. Forearm 5. Palm • Measured the following 1. Melanin 2. Hemoglobin 3. Scattering 4. Epidermal Thickness This study is still unpublished
- 56. UT Biomedical Informatics Lab Instrumentation 55/60 x 2 Source Diameter = 40 μm Ring 1 Diameter = 40 μm Ring 2 Diameter = 200 μm Ring 1 SDS = 55 μm Ring 2 SDS = 205 μm Unfortunately, data from ring 1 was unusable
- 57. UT Biomedical Informatics Lab Melanin 56/60 Back Calf Cheek Forearm Palm 0 0.5 1 1.5 2 2.5 3 3.5 4 MelaninConcentration(mg/ml) Palm has less melanin, which agrees with the expected result. Average of 1.83 mg/ml is within range of published values for melanin concentration [0-5 mg/ml]
- 58. UT Biomedical Informatics Lab Hemoglobin 57/60 Back Calf Cheek Forearm Palm 0 0.5 1 1.5 2 2.5 3 3.5 HemoglobinConcentration(mg/ml) Higher levels of hemoglobin in the face and forearm agrees with the expected results. Average of 1.37 mg/ml is within range of published values for [Hb] [0.5-10 mg/ml]
- 59. UT Biomedical Informatics Lab Scattering 58/60 No significant difference between anatomical locations. Average of 22.75 cm-1 is within range of published values for scattering at 630 nm [15 – 25 cm-1]
- 60. UT Biomedical Informatics Lab Epidermal Thickness 59/60 Back Calf Cheek Forearm Palm 0 20 40 60 80 100 120 140 160 EpidermalThickness(mm) No significant difference between anatomical locations. Average of 90 μm is within range of published values for epidermal thickness. [40 – 200 μm] We expected to see a difference between anatomical locations.
- 61. UT Biomedical Informatics Lab Conclusions about Pilot Study A two-layer model can be used to extract depth dependent properties from in vivo DRS data The results agree with previously published values However, we expected to see a difference between anatomical locations for epidermal thickness – This could be due to the absence of data from the inner ring of fibers. 60/60
- 62. UT Biomedical Informatics Lab Conclusions about Pilot Study Data should be recollected with multiple SDSs Additional patient data such as race/ethnicity and skin color should be documented 61
- 63. UT Biomedical Informatics Lab Overall Conclusions The MCLUT method is an accurate and fast way to analyze DRS data A one-layer assumption for skin causes significant errors in DRS data analysis The MCLUT method can be extended to two-layers, allowing the extraction of depth dependent properties Depth sampling of DRS probes can be tuned by changing the probe geometry DRS can be used to measure the depth dependent properties of skin in vivo 62/60
- 64. UT Biomedical Informatics Lab Contributions to Field 63 The MCLUT Method Two-Layer MCLUT Method One-Layer Errors Analysis DRS Sampling Depth Analysis
- 65. UT Biomedical Informatics Lab Funding The NIH (R21EB0115892) The NSF (DGE-1110007) CPRIT (RP130702) 64
- 66. UT Biomedical Informatics Lab Acknowledgements 65 The Biophotonics Lab • James W. Tunnell • Will Goth • Bin Yang • Manu Sharma • Sam Lim • Sheldon Bish • Xu Feng • Varun Patani The Biomedical Informatics Lab • Mia K. Markey • Nishant Verma • Gezheng Wen • Clement Sun • Nisha Kumaraswamy • Hans Huang • Juhun Lee • Gautam Muralidhar • Daifeng Wang UT BME Staff • Margo Cousins • Brittain Sobey • Michael Don

























![UT Biomedical Informatics Lab
Fit Two-Layer Data with One Layer Model
26/60
Hb + HbO2
melanin
melanin +
Hb + HbO2
1. Create two-layer
spectra
2. Fit with one-
layer model
• [mel]
• [Hb]
• SO2
• Scattering
• Epidermal thickness
• [mel]
• [Hb]
• SO2
• Scattering
• Vessel radius
Notice that the fit
is very good](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-27-320.jpg)
![UT Biomedical Informatics Lab
Melanin
27/60
• One-Layer model
underestimates [mel]
• Magnitude of error is
dependent on epidermal
thickness (Z0)
• Z0 Error](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-28-320.jpg)
![UT Biomedical Informatics Lab
Hemoglobin
28/60
• One-Layer model
underestimates [Hb]
• Magnitude of error is
dependent on epidermal
thickness (Z0)
• Z0 Error](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-29-320.jpg)



![UT Biomedical Informatics Lab
Correlation Between [Hb] and [mel]
32/60
R = 0.04 R = 0.80
One-layer assumption causes artificial correlation
between [Hb] and [mel]](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-33-320.jpg)
![UT Biomedical Informatics Lab
Conclusions about One-Layer Errors
Causes underestimation of [Hb] and [mel]
– Magnitude of error is function of epidermal
thickness
Causes error in SO2 that is a function of
epidermal thickness as well as SO2
Vessel Packaging factor and epidermal
thickness are highly correlated
Causes an artificial correlation in [Hb] and
[mel]
33/60](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-34-320.jpg)
















![UT Biomedical Informatics Lab
Analytical Model of Sampling Depth
50/60
This expression can be
used to aid in the design
of application specific
DRS probesE = 2.89%
Expression was found using
TableCurve 3D. Free parameters
[a1, a2, a3, a4] were selected using a
least-squares fitting algorithm.](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-51-320.jpg)





![UT Biomedical Informatics Lab
Melanin
56/60
Back Calf Cheek Forearm Palm
0
0.5
1
1.5
2
2.5
3
3.5
4
MelaninConcentration(mg/ml)
Palm has less melanin,
which agrees with the
expected result.
Average of 1.83 mg/ml
is within range of
published values for
melanin concentration
[0-5 mg/ml]](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-57-320.jpg)
![UT Biomedical Informatics Lab
Hemoglobin
57/60
Back Calf Cheek Forearm Palm
0
0.5
1
1.5
2
2.5
3
3.5
HemoglobinConcentration(mg/ml)
Higher levels of
hemoglobin in the face
and forearm agrees
with the expected
results.
Average of 1.37 mg/ml
is within range of
published values for
[Hb]
[0.5-10 mg/ml]](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-58-320.jpg)
![UT Biomedical Informatics Lab
Scattering
58/60
No significant
difference between
anatomical locations.
Average of 22.75 cm-1
is within range of
published values for
scattering at 630 nm
[15 – 25 cm-1]](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-59-320.jpg)
![UT Biomedical Informatics Lab
Epidermal Thickness
59/60
Back Calf Cheek Forearm Palm
0
20
40
60
80
100
120
140
160
EpidermalThickness(mm)
No significant
difference between
anatomical locations.
Average of 90 μm is
within range of
published values for
epidermal thickness.
[40 – 200 μm]
We expected to see a
difference between
anatomical locations.](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-60-320.jpg)





