BITS training - UCSC Genome Browser - Part 2
- 1. UCSC genome browsing Paco Hulpiau http://www.bits.vib.be
- 2. TABLE GET CURRENT BROWSER DNA BROWSER GRAPHIC IN PDF TO GET OTHER CLICK DATA LINE
- 3. TO GET OTHER CLICK LINE 2 DATA
- 4. Databases & accession numbers § GenBank exchanges data daily with its two partners in the International Nucleotide Sequence Database Collaboration (INSDC): European Bioinformatics Institute (EBI, part of EMBL) DNA Data Bank of Japan (DDBJ) § Characteristics of GenBank and RefSeq @ NCBI : GenBank RefSeq Curated, NCBI creates from existing Not curated, author submits data Multiple records for same loci Single records for each molecule No limit to species included Limited to model organisms
- 5. Databases & accession numbers § § The Ensembl automatic gene annotation system (Curwen et al, 2004) : The gene-building system enables fast automated annotation of eukaryotic genomes. It annotates genes based on evidence derived from known protein, cDNA, and EST sequences incl. GenBank sequences shared by INSDC, UniProtKB and NCBI RefSeq
- 6. Databases & accession numbers § Database Typical accession numbers GenBank AAA37420 NM_123456 = mRNA NP_123456 = proteins RefSeq XM_123456 = predicted mRNA XP_123456 = predicted proteins UniProtKB (Swiss- P12345, Q1AAA9 Prot/TrEMBL) ENSMUSG00000123456 for Genes Ensembl ENSMUST00000123456 for Transcripts ENSMUSP00000123456 for Proteins
- 7. TO GET OTHER CLICK LINE 2 DATA
- 27. zoom in on exon 1 + upstream
- 36. Exercises (II) 1) Are there any diseases related to your gene of interest? (OMIM) Which interactions partners are known? (Entrez Gene) Any important SNPs changing the amino acid sequence? Get the multiple sequence alignment (MSA, multiz46way) showing the nucleotide sequences of human, mouse, chicken, Xenopus and zebrafish genes (CDS fasta alignment, exons not separate). Save your results (e.g. exercises2_1.doc).
- 37. 3 GET DNA TO GET OTHER DATA
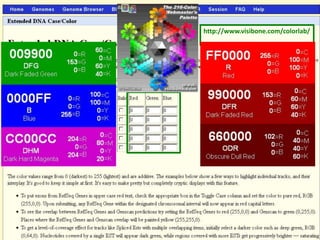
- 50. Exercises (II) 2) Get the DNA sequence for your gene of interest including 2000 base pairs upstream and use the following extended case/color options: » RefSeq and Ensembl genes in bold » SNPs (132) underlined » Regulatory information e.g. from Oreganno and miRNA sites in different colors » Save your results (e.g. exercises2_2a.doc).
- 51. Exercises (II) 2) Try to get the DNA sequence for your gene of interest in chicken or zebrafish and use the following extended case/color options: » UCSC, RefSeq and Ensembl genes in bold » Other RefSeq genes underlined » Human proteins in a specific color » Save your results (e.g. exercises2_2b.doc).
- 52. 4 TABLE BROWSER TO GET OTHER DATA
- 59. COPY (Ctrl+C)
- 65. = Accession Number (RefSeq) e.g. NM_001229 = Gene Name (Entrez) e.g. CASP1
- 71. Exercises (II) 3) Get a list of the RefSeq and Ensembl transcripts using the table browser with the following selected fields: » name, chromosome, exon count, name2 » Save the results (exercises2_3a.xls) Also get the sequences and save as genename_transcripts.fasta Search the mouse genome using the filter in the table browser to get all family members of a protein family (research interest) and save the results in a list (exercises2_3b.xls) containing name, chromosome, cds start and end, exon count and name2
- 75. BLAT = Blast-Like Alignment Tool Ø search for high similarity matches by indexing entire genome Ø DNA limit = 25000 bases, for multiple seqs 50000 bases Ø protein limit = 10000 aa, for multiple seqs 25000 aa Ø total sequences = 25
- 76. PASTE (Ctrl+V)
- 85. TTTAGCCAACGAACAGTCGCT TTCTCTTTGCATCTGTCCCAG
- 88. § The Utilities page contains links to some tools created by the UCSC Genome Bioinformatics Group. § DNA Duster & Protein Duster remove non-sequence related characters from an input sequence.
- 91. Exercises (II) 4) Use BLAT to find orthologs of your gene in chicken, zebrafish and fruit fly. What is the genomic location? Are the flanking genes the same? Perform an in silico PCR to see what happens when more than 1 PCR product may arise and determine product size and Tm: species: human forward primer: TTC AAG GAG GCC TTC TCC CT reverse primer: CTG GGG GAG AAG CTG A (+click flip reverse)