CPSC 340: Machine Learning and Data Mining More Clustering Andreas Lehrmann and Mark Schmidt University of British Columbia, Fall 2022
- 1. CPSC 340: Machine Learning and Data Mining More Clustering Andreas Lehrmann and Mark Schmidt University of British Columbia, Fall 2022 https://www.students.cs.ubc.ca/~cs-340
- 2. Last Time: K-Means Clustering • We want to cluster data: – Assign examples to groups. • K-means clustering: – Define groups by “means” – Assigns examples to nearest mean. (And updates means during training.) • Issues with k-means: – Fast but sensitive to initialization. – Choosing ‘k’ is annoying.
- 3. Vector Quantization • K-means originally comes from signal processing. • Designed for vector quantization: – Replace examples with the mean of their cluster (“prototype”). • Example: – Facebook places: 1 location summarizes many. – What sizes of clothing should I make? http://wannabite.com/wp-content/uploads/2014/10/ragu-pasta-sauce-printable-coupon.jpg
- 4. Vector Quantization for Basketball Players • Clustering NBA basketball players based on shot type/percentage: • The “prototypes” (means) give offensive styles (like “catch and shoot”). https://fansided.com/2018/08/23/nylon-calculus-shooting-volume-versus-efficiency/
- 6. (Bad) Vector Quantization in Practice • Political parties can be thought as a form of vector quantization: – Hope is that parties represent what a cluster of voters want. • With larger ‘k’ more voters have a party that closely reflects them. • With smaller ‘k’, parties are less accurate reflections of people’s views. https://globalnews.ca/news/5191123/federal-election-seat-projection-trudeau-liberals-minority/
- 7. Shape of K-Means Clusters • Recall that k-means assigns cluster based on nearest mean. • This leads to partitions the space : • Observe that the clusters are convex regions (proof in bonus).
- 8. Convex Sets • A set is convex if line between two points in the set stays in the set. Convex Convex Not Convex
- 9. Shape of K-Means Clusters Animation
- 10. K-Means with Non-Convex Clusters https://corelifesciences.com/human-long-non-coding-rna-expression-microarray-service.html
- 11. K-Means with Non-Convex Clusters https://corelifesciences.com/human-long-non-coding-rna-expression-microarray-service.html K-means cannot separate some non-convex clusters
- 12. K-Means with Non-Convex Clusters https://corelifesciences.com/human-long-non-coding-rna-expression-microarray-service.html K-means cannot separate some non-convex clusters Though over-clustering can help (“hierarchical”)
- 14. John Snow and Cholera Epidemic • John Snow’s 1854 spatial histogram of deaths from cholera: • Found cluster of cholera deaths around a particular water pump. – Went against airborne theory, but pump later found to be contaminated. – “Father” of epidemiology. https://en.wikipedia.org/wiki/John_Snow
- 15. Motivation for Density-Based Clustering • Density-based clustering: – Clusters are defined by “dense” regions. – Examples in non-dense regions don’t get clustered. • Not trying to “partition” the space. • Clusters can be non-convex: – Elephant clusters affected by vegetation, mountains, rivers, water access, etc. • It’s a non-parametric clustering method: – No fixed number of clusters ‘k’. – Clusters can become more complicated with more data. http://www.defenders.org/elephant/basic-facts
- 16. Other Potential Applications • Where are high crime regions of a city? • Where should taxis patrol? • Where can proteins ‘dock’? • Where are people tweeting? • Where does Iguodala make/miss shots? • Which products are similar to this one? • Which pictures are in the same place? https://en.wikipedia.org/wiki/Cluster_analysis https://www.flickr.com/photos/dbarefoot/420194128/ http://letsgowarriors.com/replacing-jarrett-jack/2013/10/04/ http://www.dbs.informatik.uni-muenchen.de/Forschung/KDD/Clustering/
- 17. Density-Based Clustering in Action Interactive demo
- 18. Density-Based Clustering • Density-based clustering algorithm (DBSCAN) has two hyperparameters: – Epsilon (ε): distance we use to decide if another point is a “neighbour”.
- 19. Density-Based Clustering • Density-based clustering algorithm (DBSCAN) has two hyperparameters: – Epsilon (ε): distance we use to decide if another point is a “neighbour”.
- 20. Density-Based Clustering • Density-based clustering algorithm (DBSCAN) has two hyperparameters: – Epsilon (ε): distance we use to decide if another point is a “neighbour”. – MinNeighbours: number of neighbours needed to say a region is “dense”. • If you have at least minNeighbours “neighbours”, you are called a “core” point. • Main idea: merge all neighbouring core points to form clusters.
- 25. • Intuitively, density-based clustering algorithm implements a “chain reaction” throughout the dense areas. • For each example xi: – If xi is already assigned to a cluster, do nothing. – Test whether xi is a ‘core’ point (≥ minNeighbours examples within ‘ε’). • If xi is not core point, do nothing (this could be an outlier). • If xi is a core point, make a new cluster and call the “expand cluster” function. – Which spreads the “reaction” to nearby points. Density-Based Clustering Pseudo-Code
- 26. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 27. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 28. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 29. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 30. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 31. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 32. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 33. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 34. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 35. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 36. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 37. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 38. Density-Based Clustering Pseudo-Code • “Expand cluster” function: – Assign to this cluster all xj within distance ‘ε’ of core point xi to this cluster. – For each new “core” point found, call “expand cluster” (recursively).
- 39. Density-Based Clustering Issues • Some points are not assigned to a cluster. – Good or bad, depending on the application. • Ambiguity of “non-core” (boundary) points: • Sensitive to the choice of ε and minNeighbours. – Original paper proposed an “elbow” method (see bonus slide). – Otherwise, not sensitive to initialization (except for boundary points). • If you get a new example, finding cluster is expensive. – Need to compute distances to core points (or maybe all training points). • In high-dimensions, need a lot of points to ‘fill’ the space.
- 40. Density-Based Clustering in Bacteria • Quorum sensing: – Bacteria continuously release a particular molecule. – They have sensors for this molecule. • If sensors become very active: – It means cell density is high. – Causes cascade of changes in cells. (Some cells “stick together” to form a physical cluster via “biofilm”.) https://en.wikipedia.org/wiki/Quorum_sensing
- 41. Density-Based Clustering in People • “High density leading a chain reaction” can also happen in people. – “Social distancing”: try to reduce the number of people within 𝜖. – “Wearing masks”: try to increase the 𝜖 needed for a chain reaction.
- 42. Next Topic: Ensemble Clustering
- 43. Ensemble Clustering • We can consider ensemble methods for clustering. – “Consensus clustering” • It’s a good/important idea: – Bootstrapping is widely-used. – “Do clusters change if the data was slightly different?” • But we need to be careful about how we combine models.
- 44. Ensemble Clustering • E.g., run k-means 20 times and then cluster using the mode of each ! 𝑦i. • Normally, averaging across models doing different things is good. • But this is a bad ensemble method: worse than k-means on its own.
- 45. Label Switching Problem • This doesn’t work because of “label switching” problem: – The cluster labels $ 𝑦i are meaningless. – We could get same clustering with permuted labels (“exchangeable”): – All $ 𝑦i become equally likely as number of initializations increases.
- 46. Addressing Label Switching Problem • Ensembles can’t depend on label “meaning”: – Don’t ask “is point xi in red square cluster?”, which is meaningless. – Ask “is point xi in the same cluster as xj?”, which is meaningful. – Bonus slides give an example method (“UBClustering”).
- 47. Next Topic: Hierarchical Clustering
- 48. Differing Densities • Consider density-based clustering on this data:
- 49. Differing Densities • Increase epsilon and run it again: • There may be no density-level that gives you 3 clusters.
- 50. Differing Densities • Here is a worse situation: • Now you need to choose between coarse/fine clusters. • Instead of fixed clustering, we often want hierarchical clustering.
- 51. Hierarchical Clustering • Hierarchical clustering produces a tree of clusterings. – Each node in the tree splits the data into 2 or more clusters. – Much more information than using a fixed clustering. – Often have individual data points as leaves. GIF
- 52. Application: Phylogenetics • We sequence genomes of a set of organisms. • Can we construct the “tree of life”? • Comments on this application: – On the right are individuals. – As you go left, clusters merge. – Merges are ‘common ancestors’. • More useful information in the plot: – Line lengths: chosen here to approximate time. – Numbers: #clusterings across bootstrap samples. – ‘Outgroups’ (walrus, panda) are a sanity check. http://www.nature.com/nature/journal/v438/n7069/fig_tab/nature04338_F10.html
- 53. Application: Phylogenetics • Interactive demo of model of full tree of life: www.onezoom.org
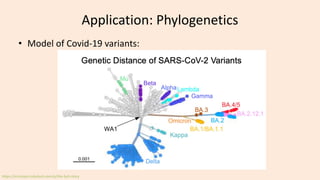
- 54. Application: Phylogenetics • Model of Covid-19 variants: https://erictopol.substack.com/p/the-ba5-story
- 55. Application: Phylogenetics • Comparative method in linguistics studies evolution of languages: https://en.wikipedia.org/wiki/Comparative_method_(linguistics)
- 56. Application: Phylogenetics • January 2016: evolution of fairy tales. – Evidence that “Devil and the Smith” goes back to bronze age. – “Beauty and the Beast” published in 1740, but might be 2500-6000 years old. http://rsos.royalsocietypublishing.org/content/3/1/150645
- 57. Application: Phylogenetics • January 2016: evolution of fairy tales. – Evidence that “Devil and the Smith” goes back to bronze age. – “Beauty and the Beast” published in 1740, but might be 2500-6000 years old. • September 2016: evolution of myths. – “Cosmic hunt” story: • Person hunts animal that becomes constellation. – Previously known to be at least 15,000 years old. • May go back to paleololithic period. http://www.nature.com/nature/journal/v438/n7069/fig_tab/nature04338_F10.html
- 58. Application: Fashion? • Hierarchical clustering of clothing material words in Vogue: http://dh.library.yale.edu/projects/vogue/fabricspace/
- 59. Agglomerative (Bottom-Up) Clustering • Most common hierarchical method: agglomerative clustering. 1. Starts with each point in its own cluster. https://en.wikipedia.org/wiki/Hierarchical_clustering
- 60. Agglomerative (Bottom-Up) Clustering • Most common hierarchical method: agglomerative clustering. 1. Starts with each point in its own cluster. 2. Each step merges the two “closest” clusters. https://en.wikipedia.org/wiki/Hierarchical_clustering
- 61. Agglomerative (Bottom-Up) Clustering • Most common hierarchical method: agglomerative clustering. 1. Starts with each point in its own cluster. 2. Each step merges the two “closest” clusters. https://en.wikipedia.org/wiki/Hierarchical_clustering
- 62. Agglomerative (Bottom-Up) Clustering • Most common hierarchical method: agglomerative clustering. 1. Starts with each point in its own cluster. 2. Each step merges the two “closest” clusters. https://en.wikipedia.org/wiki/Hierarchical_clustering
- 63. Agglomerative (Bottom-Up) Clustering • Most common hierarchical method: agglomerative clustering. 1. Starts with each point in its own cluster. 2. Each step merges the two “closest” clusters. https://en.wikipedia.org/wiki/Hierarchical_clustering
- 64. Agglomerative (Bottom-Up) Clustering • Most common hierarchical method: agglomerative clustering. 1. Starts with each point in its own cluster. 2. Each step merges the two “closest” clusters. 3. Stop with one big cluster that has all points. https://en.wikipedia.org/wiki/Hierarchical_clustering Animation
- 65. Agglomerative (Bottom-Up) Clustering • Reinvented by different fields under different names (“UPGMA”). • Needs a “distance” between two clusters. • A standard choice: distance between means of the clusters. – Not necessarily the best, many choices exist (bonus slide). • Cost is O(n3d) for basic implementation. – Each step costs O(n2d), and each step might only cluster 1 new point.
- 66. Summary • Shape of K-means clusters: – Partitions space into convex sets. • Density-based clustering: – “Expand” and “merge” dense regions of points to find clusters. – Not sensitive to initialization or outliers. – Useful for finding non-convex connected clusters. • Ensemble clustering: combines multiple clusterings. – Can work well but need to account for label switching. • Hierarchical clustering: more informative than fixed clustering. • Agglomerative clustering: standard hierarchical clustering method. – Each point starts as a cluster, sequentially merge clusters. • Next time: • Discovering (and then ignoring) a hole in the ozone layer.
- 67. Why are k-means clusters convex? • K-means clusters are formed by the intersection of half-spaces. Half-space
- 68. Why are k-means clusters convex? • K-means clusters are formed by the intersection of half-spaces. Half-space Intersection Half-space
- 69. Why are k-means clusters convex?
- 70. Why are k-means clusters convex? “Closer to red” half-space “Closer to green” half-space
- 71. Why are k-means clusters convex? “Closer to red” half-space “Closer to green” half-space
- 72. Why are k-means clusters convex? Blue over green half-space Green over blue half-space
- 73. Why are k-means clusters convex? Magenta over green half-space Green over magenta half-space
- 74. Why are k-means clusters convex?
- 75. Why are k-means clusters convex? • Half-spaces are convex sets. • Intersection of convex sets is a convex set. – Line segment between points in each set are still in each set. • So intersection of half-spaces is convex. Half-space Intersection Half-space
- 76. Why are k-means clusters convex? • Formal proof that “cluster 1” is convex (works for other clusters).
- 77. Voronoi Diagrams • The k-means partition can be visualized as a Voronoi diagram: • Can be a useful visualization of “nearest available” problems. – E.g., nearest tube station in London. http://datagenetics.com/blog/may12017/index.html
- 79. “Elbow” Method for Density-Based Clustering • From the original DBSCAN paper: – Choose some ‘k’ (they suggest 4) and set minNeighbours=k. – Compute distance of each points to its ‘k’ nearest neighbours. – Sort the points based on these distances and plot the distances: – Look for an “elbow” to choose 𝜖. https://www.aaai.org/Papers/KDD/1996/KDD96-037.pdf
- 80. OPTICS • Related to the DBSCAN “elbow” is “OPTICS”. – Sort the points so that neighbours are close to each other in the ordering. – Plot the distance from each point to the next point. – Clusters should correspond to sequencers with low distance. https://en.wikipedia.org/wiki/OPTICS_algorithm
- 81. UBClustering Algorithm • Let’s define a new ensemble clustering method: UBClustering. 1. Run k-means with ‘m’ different random initializations. 2. For each example i and j: – Count the number of times xi and xj are in the same cluster. – Define p(i,j) = count(xi in same cluster as xj)/m. 3. Put xi and xj in the same cluster if p(i,j) > 0.5. • Like DBSCAN merge clusters in step 3 if i or j are already assigned. – You can implement this with a DBSCAN code (just changes “distance”). – Each xi has an xj in its cluster with p(i,j) > 0.5. – Some points are not assigned to any cluster.
- 83. Distances between Clusters • Other choices of the distance between two clusters: – “Single-link”: minimum distance between points in clusters. – “Average-link”: average distance between points in clusters. – “Complete-link”: maximum distance between points in clusters. – Ward’s method: minimize within-cluster variance. – “Centroid-link”: distance between a representative point in the cluster. • Useful for distance measures on non-Euclidean spaces (like Jaccard similarity). • “Centroid” often defined as point in cluster minimizing average distance to other points.
- 84. Cost of Agglomerative Clustering • One step of agglomerative clustering costs O(n2d): – We need to do the O(d) distance calculation between up to O(n2) points. – This is assuming the standard distance functions. • We do at most O(n) steps: – Starting with ‘n’ clusters and merging 2 clusters on each step, after O(n) steps we’ll only have 1 cluster left (though typically it will be much smaller). • This gives a total cost of O(n3d). • This can be reduced to O(n2d log n) with a priority queue: – Store distances in a sorted order, only update the distances that change. • For single- and complete-linkage, you can get it down to O(n2d). – “SLINK” and “CLINK” algorithms.
- 85. Bonus Slide: Divisive (Top-Down) Clustering • Start with all examples in one cluster, then start dividing. • E.g., run k-means on a cluster, then run again on resulting clusters. – A clustering analogue of decision tree learning.