ENZYME BIOCHEMISTRY
- 1. ENZYME-BIOCHEMISTRY Dr. NARESH PANIGRAHI ASST.PROFESSOR GITAM INSTITUTE OF PHARMACY GITAM UNIVERSITY VISAKHAPATNAM
- 2. ENZYMES • Enzymes are the body's catalysts. • Without them, the cell's chemical reactions would be too slow and many would not occur at all. • A catalyst is an agent which speeds up a chemical reaction without being changed itself. • As an example, let us consider the substrate for lactate dehydrogenase—an enzyme which catalyses the reduction of pyruvic acid to lactic acid
- 3. • They increase the rate of a reaction by lowering the activation energy barrier.
- 4. How do catalysts lower activation energies? • There are several factors at work. Catalysts provide a reaction surface or environment. Catalysts bring reactants together. Catalysts position reactants correctly so that they easily attain their transition state configurations. Catalysts weaken bonds. Catalysts may participate in the mechanism.
- 5. The Mechanism of Enzymatic Action
- 6. The reactant usually binds to the enzyme forming a transition complex that stabilizes the transition state. • The interaction between enzyme and substrate is very specific. Specificity may be for a class of compounds or for a particular compound. Eg. Hexokinase / glucokinase
- 7. Enzyme Structure • All known enzymes are proteins. • They are high molecular weight compounds made up principally of chains of amino acids linked together by peptide bonds. • Many enzymes require the presence of cofactors • Coenzyme= a non-protein organic substance which is loosely attached to the protein part.(NAD+, NADH, FAD) • Prosthetic group = an organic substance which is firmly attached to the protein or apoenzyme portion or • Metal-ion activators ( K+, Fe++, Fe+++, Cu++, Co++, Zn++, Mn++, Mg++, Ca++)
- 9. The structure of an ENZYME CONTAINS • AN Apoenzyme: Protein part • Cofactor: Non-protein component – Coenzyme: Organic cofactor • Holoenzyme: Apoenzyme plus cofactor
- 10. Specificity of Enzymes • One of the properties of enzymes that makes them so important as diagnostic and research tools is the specificity they exhibit relative to the reactions they catalyze. • In general, there are four distinct types of specificity: 1. Absolute specificity - the enzyme will catalyze only one reaction. 2. Group specificity - the enzyme will act only on molecules that have specific functional groups, such as amino, phosphate and methyl groups. 3. Linkage specificity - the enzyme will act on a particular type of chemical bond regardless of the rest of the molecular structure. 4. Stereochemical specificity - the enzyme will act on a particular steric or optical isomer.
- 11. • In the early days, the enzymes were given names by their discoverers in an arbitrary manner. For example, the names pepsin, trypsin and chymotrypsin convey no information about the function of the enzyme or the nature of the substrate on which they act. • Sometimes, the suffix-ase was added to the substrate for naming the enzymes e.g. lipase acts on lipids; nuclease on nucleic acids; lactase on lactose. • These are known as trivial names of the enzymes which, however, fail to give complete information of enzyme reaction (type of reaction, cofactor requirement etc.) Nomenclature and Enzyme Classification
- 14. Enzyme nomenclature EC nomenclature * each enzyme is classified by EC number (Enzyme Commission of IUBMB) – 6 classes: • EC 1.x.x.x oxidoreductases • EC 2.x.x.x transferases • EC 3.x.x.x hydrolases • EC 4.x.x.x lyases • EC 5.x.x.x isomerases • EC 6.x.x.x ligases (synthetases) → classification by a reaction catalyzed by the enzyme
- 15. systematic names * are made according to a special rules, they specify a reaction catalyzed by the enzyme example: ATP : D-glucose phosphotransferase (EC 2.7.1.2) transfers (2) phosphate (7) to an alcohol group (1) ATP + D-Glc ADP + D-Glc-6-phosphate (Glc-6-P)
- 16. common names (= accepted names) * easier than the systematic names, widely used * very important! example: EC 2.7.1.2. = glucokinase (see above)
- 17. Nomenclature and Enzyme Classification • There are 6 main classes based on the type of reaction catalyzed. • Oxidoreductase: Oxidation-reduction reactions • Transferase: Transfer functional groups • Hydrolase: Hydrolysis • Lyase: Removal of atoms without hydrolysis • Isomerase: Rearrangement of atoms • Ligase: Joining of molecules, uses ATP
- 26. THE ACTIVE SITE OF AN ENZYME • ACTIVE SITE: IS a Pocket in the enzyme where substrates bind and catalytic reaction occurs. • Active site is a relatively small 3-D region within the enzyme. • Substrates bind in active site by weak non- covalent interactions • A. hydrogen bonding • B. hydrophobic interactions • C. ionic interactions
- 27. THE ACTIVE SITE OF AN ENZYME • The amino acids present in the active site play an important role in enzyme function. • Amino acids present in the active site can have one of two roles. 1. Binding—the amino acid residue is involved in binding the substrate to the active site. 2. Catalytic—the amino acid is involved in the mechanism of the reaction.
- 30. THEORIES OF ENZYME ACTION
- 33. Allosteric Enzymes • Allosteric enzymes have one or more allosteric sites • Allosteric sites are binding sites distinct from an enzyme’s active site or substrate-binding site • Molecules that bind to allosteric sites are called effectors or modulators
- 34. Binding to allosteric sites alters the activity of the enzyme. This is called cooperative binding. Effectors may be positive or negative Effectors may be homotropic or heterotropic Regulatory enzymes of metabolic pathways are allosteric enzymes (eg: feedback inhibition)
- 35. Factors affecting Enzymes • substrate concentration • pH • temperature • inhibitors © 2007 Paul Billiet ODWS
- 36. Substrate concentration: Non-enzymic reactions • The increase in velocity is proportional to the substrate concentration Reaction velocity Substrate concentration © 2007 Paul Billiet ODWS
- 37. Substrate concentration: Enzymic reactions • Faster reaction but it reaches a saturation point when all the enzyme molecules are occupied. • If you alter the concentration of the enzyme then Vmax will change too. Reaction velocity Substrate concentration Vmax © 2007 Paul Billiet ODWS
- 38. The effect of pH Optimum pH values Enzyme activity Trypsin Pepsin pH 1 3 5 7 9 11 © 2007 Paul Billiet ODWS
- 39. The effect of pH • Extreme pH levels will produce denaturation • The structure of the enzyme is changed • The active site is distorted and the substrate molecules will no longer fit in it • At pH values slightly different from the enzyme’s optimum value, small changes in the charges of the enzyme and it’s substrate molecules will occur • This change in ionisation will affect the binding of the substrate with the active site. © 2007 Paul Billiet ODWS
- 40. The effect of temperature • Q10 (the temperature coefficient) = the increase in reaction rate with a 10°C rise in temperature. • For chemical reactions the Q10 = 2 to 3 (the rate of the reaction doubles or triples with every 10°C rise in temperature) • Enzyme-controlled reactions follow this rule as they are chemical reactions • BUT at high temperatures proteins denature • The optimum temperature for an enzyme controlled reaction will be a balance between the Q10 and denaturation. © 2007 Paul Billiet ODWS
- 41. The effect of temperature Temperature / °C Enzyme activity 0 10 20 30 40 50 Q10 Denaturation © 2007 Paul Billiet ODWS
- 42. The effect of temperature • For most enzymes the optimum temperature is about 30°C • Many are a lot lower, cold water fish will die at 30°C because their enzymes denature • A few bacteria have enzymes that can withstand very high temperatures up to 100°C • Most enzymes however are fully denatured at 70°C © 2007 Paul Billiet ODWS
- 43. Inhibitors • Inhibitors are chemicals that reduce the rate of enzymic reactions. • The are usually specific and they work at low concentrations. • They block the enzyme but they do not usually destroy it. • Many drugs and poisons are inhibitors of enzymes in the nervous system. © 2007 Paul Billiet ODWS
- 44. The effect of enzyme inhibition • Irreversible inhibitors: Combine with the functional groups of the amino acids in the active site, irreversibly. Examples: nerve gases and pesticides, containing organophosphorus, combine with serine residues in the enzyme acetylcholine esterase. © 2007 Paul Billiet ODWS
- 45. The effect of enzyme inhibition • Reversible inhibitors: These can be washed out of the solution of enzyme by dialysis. There are two categories. © 2007 Paul Billiet ODWS
- 46. The effect of enzyme inhibition 1. Competitive: These compete with the substrate molecules for the active site. The inhibitor’s action is proportional to its concentration. Resembles the substrate’s structure closely. Enzyme inhibitor complex Reversible reaction E + I EI © 2007 Paul Billiet ODWS
- 47. The effect of enzyme inhibition Succinate Fumarate + 2H++ 2e- Succinate dehydrogenase CH2COOH CH2COOH CHCOOH CHCOOH COOH COOH CH2 Malonate © 2007 Paul Billiet ODWS
- 49. Example
- 50. The effect of enzyme inhibition 2. Non-competitive: These are not influenced by the concentration of the substrate. It inhibits by binding irreversibly to the enzyme but not at the active site. Examples • Cyanide combines with the Iron in the enzymes cytochrome oxidase. • Heavy metals, Ag or Hg, combine with –SH groups. These can be removed by using a chelating agent such as EDTA. © 2007 Paul Billiet ODWS
- 51. Applications of inhibitors • Negative feedback: end point or end product inhibition • Poisons snake bite, plant alkaloids and nerve gases. • Medicine antibiotics, sulphonamides, sedatives and stimulants © 2007 Paul Billiet ODWS
- 53. Enzyme inhibitors in Medicine • The effectiveness of an enzyme inhibitor as a therapeutic agent will depend on (1) The potency of the inhibitor, (2) Its specificity toward its target enzyme, (3)The choice of metabolic pathway targeted for disruption, and (4) The inhibitor or a derivative possessing appropriate pharmacokinetic characteristics.
- 57. Coenzyme • The protein part of the enzyme, on its own, is not always adequate to bring about the catalytic activity. • Many enzymes require certain non-protein small additional factors, collectively referred to as cofactors for catalysis. • The cofactors may be organic or inorganic in nature.
- 58. The structure of an ENZYME CONTAINS • AN Apoenzyme: Protein part • Cofactor: Non-protein component – Coenzyme: Organic cofactor • Holoenzyme: Apoenzyme plus cofactor
- 59. Cofactor • 1. Prosthetic group (when cofactor is very tightly bound to the apoenzyme and has small size ) • 2. Metal ion • 3. Coenzyme(organic molecule derived from the vitamin-B which participate directly in enzymatic reactions)
- 60. Defination • The non-protein, organic, Iow molecular weight and dialysable substance associated with enzyme function is known as coenzyme. • The functional enzyme is referred to as holoenzyme which is made up of a protein part (apoenzyme) and a non-protein part (coenzyme); • The term prosthetic group is used when a non- protein moiety is tightly bound to the enzyme which is not easily separable by dialysis. • The term activator is referred to the inorganic cofactor (like Ca2+, Mg2+, Mn2+ etc.; necessary to enhance enzyme activity
- 61. Functions of coenzymes • Function of apoenzyme: • It is responsible for the reaction • Function of cofactor: • It is responsible for the bonds formation between enzyme and substrate • Transfer of functional groups • Takes place in the formation of tertiary structure of protein part. • Coenzymes participate in various reactions involving transfer of atoms or groups like hydrogen, aldehyde, keto, amino, acyl, methyl, carbon dioxide etc. • Coenzymes play a decisive role in enzyme function
- 62. Prosthetic group • 1. Heme group of cytochromes • 2. Biothin group of acetyl-CoA carboxylase
- 63. Metal ions • Fe - cytochrome oxidase, catalase • Cu - cytochrome oxidase, catalase • Zn - alcohol dehydrogenase • Mg - hexokinase, glucose-6-phosphatase • K, Mg - pyruvate kinase • Na, K – ATP-ase
- 64. Coenzyme • B1 • TPP- Thiamine Pyro Phosphate • B2 • FAD- Flavin Adenine Dinucleotide • FMN- Flavin Mono Nucleotide • Pantothenic acid • Coenzyme A (CoA) • B5 • NAD – Nicotinamide Adenine Dinucleotide • NADP- Nicotinamide Adenine Dinucleotide Phosphate
- 72. Nucleotide coenzymes Some of the coenzymes possess nitrogenous base, sugars and phosphate. Such coenzymes are, therefore, regarded as nucleotides e.g. NAD+, NADP+, FMN, FAD, coenzyme A, UDPC etc
- 75. Coenzymes do not decide enzyme specificity • A particular coenzyme may participate in catalytic reactions along with different enzymes. • For instance, NAD+ acts as a coenzyme for lactate dehydrogenase and alcohol dehydrogenase. • In both the enzymatic reactions, NAD+ is involved in hydrogen transfer. • The specificity of the enzyme is mostly dependent on the apoenzyme and not on the coenzyme.
- 76. Applications of enzymes • Certain enzymes are useful as therapeutic agents, analytical reagents, manipulations in genetic and for industrial application.
- 81. Isoenzymes (isozymes) • Isoenzymes (isozymes) are enzymes which catalyze the same reaction but differ in their primary structure and physico chemical properties. • which include the structure, electrophoretic and immunological properties, • Km and Vmax values, pH optimum, • Relative susceptibility to inhibitors and • degree of denaturation.
- 82. • Isoenzymes are • produced by different genes (= true isozymes) • or produced by different post translational modification (= isoforms) • found in different compartments of a cell • found in different tissues of an organism • can be oligomers of various subunits (monomers)
- 83. Explanation for the existence of isoenzymes • Many possible reasons are offered to explain the presence of isoenzymes in the living systems. 1. lsoenzymes synthesized from different genes e.g. malate dehydrogenase of cytosol is different from that found in mitochondria. 2. Oligomeric enzymes consisting of more than one type of subunits e.g. lactate dehydrogenase and creatine phosphokinase.
- 84. 3. An enzyme may be active as monomer or oligomer e.g. glutamate dehydrogenase. 4. In glycoprotein enzymes, differences in carbohydrate content may be responsible for isoenzymese .g. alkaline phosphatase.
- 85. S. No Property E.g. 1 Electrophoretic mobility Isoenzymes of Lactate dehydrogenase have different electrophoretic mobility 2 Heat stability Alkaline phosphatase isoenzymes are either heat labile or stable 3 Inhibitor An inhibitor can inhibit only one isoenzyme of an enzyme eg. Acid phosphatase 4 Km Glucokinase and hexokinase 5 Cofactors Mitochondrial isocitrate dehydrogenase requires NAD+ , cytosolic form requires NADP+ 6 Tissue localisation LDH 1 is present in heart, LDH 5 in muscle 7 Antibodies For creatine kinase, each isoenzyme can be bound only by a specific antibody
- 86. Uses of isoenzymes • Some enzymes are specific to particular tissue. • Normally they are not seen in blood or seen in negligible levels. • Usually when a tissue dies, it gets necrosed and the enzymes inside the cells of the tissues gets released in the circulation • Measuring the levels of those enzymes aids in identification of the organ that got damaged.
- 87. lsoenzymes of lactate dehydrogenase (LDH) • LDH whose systematic name is L-lactate- NAD+ oxidoreductase (E.C. 1 .'1.1.27) catalyses the interconversion of lactate and pyruvate as shown below
- 88. Lactate dehydrogenase • It occurs in 5 possible forms in the blood serum: • LDH1 • LDH2 • LDH3 • LDH4 • LDH5 They can be separated by electrophoresis (cellulose or starch gel or agarose gel). LDHI has more positive charge and fastest in electrophoretic mobility while LDH5 is the slowest.
- 89. Structure of LDH isoenzymes • LDH is an oligomeric (tetrameric) enzyme made up of four polypeptide subunits. • Two types of subunits namely M (for muscle) and H (for heart) are produced by different genes. • M-subunit is basic while H subunit is, acidic. • The isoenzymes contain either one or both the subunits giving LDHI to LDH5. The characteristic features of LDH isoenzymes are given as below
- 90. Structure of LDH • Each contains 4 polypeptide chains which are of 2 types: A and B which are usually called M (muscle) and H (heart). • LDH1 –H H H H • LDH2 – H H H M • LDH3 – H H M M • LDH4 – H M M M • LDH5 – M M M M
- 92. Significance Of Differential Catalytic Activity : • LDHl (Ha) is predominantly found in heart muscle and is inhibited by pyruvate – the substrate. Hence, pyruvate is not converted to lactate in cardiac muscle but is converted to acetyl CoA which enters citric acid cycle. • LDH5 (M+) is mostly present in skeletal muscle and the inhibition of this enzyme by pyruvate is minimal , hence pyruvate is converted to lactate. • Further, LDH5 has low K. (high affinity) while LDHl has high Km (low affinity) for pyruvate. • The differential catalytic ativities of LDHl and LDH5 in heart and skeletal muscle, respectively, are well suited for the aerobic (presence of oxygen) and anaerobic (absence of oxygen) conditions, prevailing in these tissues.
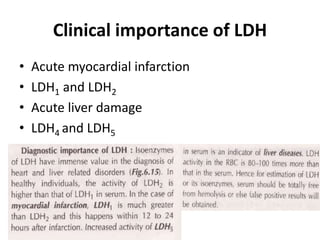
- 93. Clinical importance of LDH • Acute myocardial infarction • LDH1 and LDH2 • Acute liver damage • LDH4 and LDH5
- 97. Thank you