ENZYMES IN RECOMBINANT DNA TECHNOLOGY
- 1. DNA MODIFYING ENZYMES IN RECOMBINANT DNA TECHNOLOGY Dr. M. THIPPESWAMY
- 2. Applications • Random primer labeling of DNA. • DNA sequencing. • Second-strand cDNA synthesis. • Strand displacement amplification. DNA polymerase is an enzyme that synthesizes DNA molecules from deoxyribonucleotides, the building blocks of DNA. These enzymes are essential for DNA replication DNA polymerase
- 4. Domain organization of Klenow fragment DNA polymerase I
- 5. DNA polymerase III: The replisome is composed of the following: 2 DNA Pol III enzymes, each comprising α, ε and θ subunits. (It has been proven that there is a third copy of Pol III at the replisome.) • the α subunit (encoded by the dnaE gene) has the polymerase activity. • the ε subunit (dnaQ) has 3'→5' exonuclease activity. • the θ subunit (holE) stimulates the ε subunit's proofreading. 2 β units (dnaN) which act as slidingDNA clamps, they keep the polymerase bound to the DNA. 2 τ units (dnaX) which acts to dimerize two of the core enzymes (α, ε, and θ subunits). 1 γ unit (also dnaX) which acts as a clamp loader for the lagging strand Okazaki fragments, helping the two β subunits to form a unit and bind to DNA. The γ unit is made up of 5 γ subunits which include 3 γ subunits, 1 δ subunit (holA), and 1 δ' subunit (holB). The δ is involved in copying of the lagging strand. Χ (holC) and Ψ (holD) which form a 1:1 complex and bind to γ or τ. X can also mediate the switch from RNA primer to DNA.
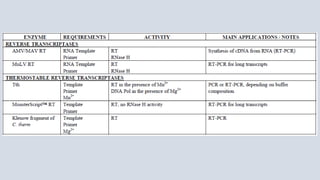
- 7. • A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. REVERSE TRANSCRIPTASE FOR MOLECULAR BIOLOGY AMV/ MAV RT • The Avian Myeloblastosis Virus (AMV) Reverse Transcriptase is an RNA-directed DNA polymerase. This enzyme can synthesize a complementary DNA strand initiating from a primer using RNA (cDNA synthesis) or single- stranded DNA as a template. • AMV/MAV RT is widely used to copy total messenger RNAs using polydT or random primers. Applications: First- and second-strand synthesis of cDNA. Primer extensions and RNA sequencing.
- 8. M-MuLV - Reverse Transcriptase • Moloney murine leukemia virus (M-MuLV) Reverse Transcriptase is a recombinant RNA-dependent DNA polymerase that uses single-stranded RNA as a template in the presence of a primer to synthesize a complementary DNA (cDNA) strand. • It can be used for the preparation of cDNA libraries, for first-strand synthesis in RT-PCR, or in dideoxy sequencing reactions in place of Klenow enzyme, for G+C–rich regions. • MuLV Reverse Transcriptase lacks endonuclease activity and has extremely low RNase H activity compared to AMV reverse transcriptase. Applications Reverse Transcriptase M-MuLV (from Moloney Murine Leukemia Virus) is used for: - In vitro synthesis of cDNA transcripts of specific RNA sequences - Preparation of cDNA libraries - Synthesis of first-strand cDNA for use in subsequent amplification reactions (RT-PCR) - Dideoxy-sequencing reactions in place of Klenow enzyme. Reverse Transcriptase M-MuLV will often synthesize through GC-rich regions where Klenow enzyme is hindered - Transcription of abundant, simple RNA target.
- 10. • Phosphatase catalyses the cleavage of a phosphate (PO4-2) group from substrate by using a water molecule (hydrolytic cleavage). This reaction is not reversible. Phosphatase Schematic representation of hydrolytic cleavage of phosphate group (-PO4-2). • On the basis of their activity there are two types of phosphatase i.e acid phosphatase and alkaline phosphatase. In both forms the alkaline phosphatase are most common. Acid phosphatase: • It shows its optimal activity at pH between 3 and 6, e.g. a lysosomal enzyme that hydrolyze organic phosphates liberating one or more phosphate groups. They are found in prostatic epithelial cells, erythrocyte, prostatic tissue, spleen, kidney etc.
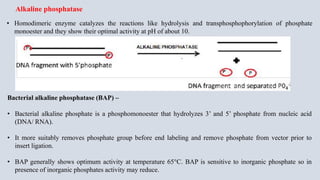
- 11. • Homodimeric enzyme catalyzes the reactions like hydrolysis and transphosphophorylation of phosphate monoester and they show their optimal activity at pH of about 10. Alkaline phosphatase Bacterial alkaline phosphatase (BAP) – • Bacterial alkaline phosphate is a phosphomonoester that hydrolyzes 3’ and 5’ phosphate from nucleic acid (DNA/ RNA). • It more suitably removes phosphate group before end labeling and remove phosphate from vector prior to insert ligation. • BAP generally shows optimum activity at temperature 65°C. BAP is sensitive to inorganic phosphate so in presence of inorganic phosphates activity may reduce.
- 12. Calf intestinal alkaline phosphatase (CIP) – It is isolated from calf intestine, which catalyzes the removal of phosphate group from 5’ end of DNA as well as RNA. This enzyme is highly used in gene cloning experiments, as to make a construct that could not undergo self- ligation. Hence after the treatment with CIP, without having a phosphate group at 5’ ends a vector cannot self ligate and recircularise. This step improves the efficiency of vector containing desired insert. Shrimp alkaline phosphatase (SAP) - Shrimp alkaline phosphatase is highly specific, heat labile phosphatase enzyme isolated from arctic shrimp (Pandalus borealis). It removes 5’ phosphate group from DNA, RNA, dNTPs and proteins. SAP has similar specificity as CIP but unlike CIP, it can be irreversibly inactivated by heat treatment at 65°C for 15mins. SAP is used for 5’ dephosphorylation during cloning experiments for various application as follows: Dephosphorylate 5’-phosphate group of DNA/RNA for subsequent labeling of the ends. To prevent self-ligation of the linearized plasmid. To prepare PCR product for sequencing. To inactivate remaining dNTPs from PCR product (for downstream sequencing appication).
- 13. Two primary uses for alkaline phosphatase in DNA modification: • Removing 5’ phosphate from different vector like plasmid, bacteriophage after treating with restriction enzyme. • This treatment prevents self ligation because unavailability of phosphate group at end. So, this treatment greatly enhances the ligation of desired insert. • During ligation of desired insert, the complementary ends of the insert and vector will come to proximity of each other (only for sticky ends but not for blunt ends). • One strand of the insert having 5’-phosphate will ligate with the 3’OH of the vector and the remaining strand will have a nick. • This nick will be sealed in the next step by ligase enzyme in the presence of ATP. • It is used to remove 5’ phosphate from fragment of DNA prior to labeling with radioactive phosphate.
- 14. • The reversible phosphorylation of proteins, catalyzed by protein kinases and protein phosphatases, is a major mechanism for the regulation of many eukaryotic cellular processes, including metabolism, muscle contraction, pre-mRNA splicing and cell cycle control. • The eukaryotic protein phosphatases comprise several families of enzymes that catalyze the dephosphorylation of intracellular phosphoproteins. • The protein phosphatases responsible for dephosphorylation of serine and threonine residues in the cytoplasmic and nuclear compartments of eukaryotic cells. • Protein phosphatases originated from two ancestor genes, one serving as the prototype for the phosphotyrosine phosphatases family (the PTP family), and the other for phosphoserine/phosphothreonine phosphatases (the PPP and PPM family). Protein phosphatases
- 15. T4 Polynucleotide kinase • T4 Polynucleotide Kinase catalyzes the transfer of the γ-phosphate from ATP to the 5´-hydroxyl terminus of double and single-stranded RNA and DNA, oligonucleotides or nucleoside 3´-monophosphates. • The enzyme is also capable of catalyzing the removal of 3´-phosphoryl groups from 3´-phosphoryl polynucleotides, deoxynucleoside 3´-monophosphates and deoxynucleoside 3´-diphosphates. • T4 PNK is widely used to end-label short oligonucleotide probes, DNA and RNA molecules.
- 16. Protein Kinase and Phosphatase reactions
- 17. PNK carries out two types of enzymatic activity: • Forward reaction: γ-phosphate is transferred from ATP to the 5' end of a polynucleotide (DNA or RNA). • Exchange reaction: target DNA or RNA having a 5' phosphate is incubated with an excess of ADP - where PNK transfers the phosphate from the nucleic acid to an ADP, forming ATP. • PNK then performs a forward reaction and transfer a phosphate from ATP to the target nucleic acid. Exchange reaction is used to label with radioactive phosphate group.
- 18. • T4 Pnkp is a homotetramer of a 301-aa (33-kDa) polypeptide, which consists of an N-terminal kinase domain of the P-loop phosphotransferase superfamily and a C-terminal phosphatase domain of the DxD acylphosphatase superfamily. • The homotetramer is formed via pairs of phosphatase-phosphatase and kinase-kinase homodimer interfaces. • Phosphate transfer activity is optimum at 37°C, pH 7.6, Mg2+ and reducing reagents (DTT or β-mercaptoethanol), and 1 mM ATP and a 5:1 ratio of ATP over 5´-OH ends. T4 Polynucleotide kinase
- 19. Applications • Labelling 5’-termini of DNA or RNA to be used as: - primers for DNA sequencing - primers for PCR - probes for hybridization - probes for transcript mapping - markers for gel electrophoresis • Addition of 5’-phosphates to oligonucleotides, PCR products, and DNA or RNA prior to ligation • Removal of 3´-phosphoryl groups • In addition to the kinase activity, T4 polynucleotide kinase also exhibits a 3´-phosphatase activity. • Optimum pH for 3’ phosphatase activity between 5 and 6, i.e. more acidic than for phosphate transfer activity. • Based on this property, protocols using T4 polynucleotide kinase as specific 3´-phosphatase have been developed (Cameron 1978). • The mutant polynucleotide kinase is very useful when the 3´-exonuclease activity must be avoided (e.g. 5´-32P terminal labeling of 3´-CMP in view of 3’ end-labeling of RNA species prior to fingerprinting or sequencing).
- 21. Terminal transferase (TdT) is a template independent polymerase that catalyzes the addition of deoxynucleotides to the 3' hydroxyl terminus of DNA molecules. Protruding, recessed or blunt-ended double or single- stranded DNA molecules serve as a substrate for TdT. The 58.3 kDa enzyme does not have 5' or 3' exonuclease activity. The addition of Co2+ in the reacton makes tailing more efficient. Terminal nucleotidyl transferase (TdT) Applications Labeling of the 3′ ends of DNA with modified nucleotides (e.g., ddNTP, DIG-dUTP) Isolated from a recombinant source Supplied with 10X Reaction Buffer Tested for the absence of endonucleases and exonucleases
- 22. Methyltransferases are a large group of enzymes that all methylate their substrates Class I Methyltransferases, all of which contain a Rossman fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, Class III methyltransferases, which are membrane associated. Methyltransferases Methyltransferases can also used in protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases.
- 23. Methyltransferase can be classified in three groups: a) m6A-generates N6 methyladenosine, b) m4C-generates N4 methylcytosine, c) m5C-generatesN5 methylcytosine. m6A and m4C methyltransferase are primarily found in prokaryotes. Restriction enzyme EcoRI cleaves within the recognition sequence if the DNA is unmethylated. On methylation by methylases, the restriction enzyme EcoRI is inhibited from cleaving within the restriction site.
- 24. THANK YOU