SQL, noSQL or no database at all? Are databases still a core skill?
- 1. SQL, noSQL or no database at all? Are databases still a core skill? Neil Saunders COMPUTATIONAL INFORMATICS www.csiro.au
- 2. Databases: Slide 2 of 24 alternative title: should David Lovell learn databases?
- 3. Databases: Slide 3 of 24 actual recent email request Hi Neil, I was wondering if you could help me with something. I am trying to put together a table but it is rather slow by hand. Do you know if you can help me with this task with a script? If it is too much of your time, don’t worry about it. Just thought I’d ask before I start. The task is: The targets listed in A tab need to be found in B tab then the entire row copied into C tab. Then the details in column C of C tab then need to be matched with the details in D tab so that the patients with the mutations are listed in row AG and AH of C tab. Again, if this isn’t an easy task for you then don’t worry about it.
- 4. Databases: Slide 4 of 24 sounds like a database to me (c. 2004)
- 5. Databases: Slide 5 of 24 database design is a profession in itself -- KEGG_DB schema CREATE TABLE ec2go ( ec_no VARCHAR(16) NOT NULL, -- EC number (with "EC:" prefix) go_id CHAR(10) NOT NULL -- GO ID ); CREATE TABLE pathway2gene ( pathway_id CHAR(8) NOT NULL, -- KEGG pathway long ID gene_id VARCHAR(20) NOT NULL -- Entrez Gene or ORF ID ); CREATE TABLE pathway2name ( path_id CHAR(5) NOT NULL UNIQUE, -- KEGG pathway short ID path_name VARCHAR(80) NOT NULL UNIQUE -- KEGG pathway name ); -- Indexes. CREATE INDEX Ipathway2gene ON pathway2gene (gene_id);
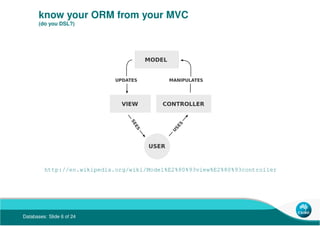
- 6. Databases: Slide 6 of 24 know your ORM from your MVC (do you DSL?) http://en.wikipedia.org/wiki/Model%E2%80%93view%E2%80%93controller
- 7. Databases: Slide 7 of 24 my one tip for today: use ORM = object relational mapping #!/usr/bin/ruby require ’sequel’ # connect to UCSC Genomes MySQL server DB = Sequel.connect(:adapter => "mysql", :host => "genome-mysql.cse.ucsc.edu", :user => "genome", :database => "hg19") # instead of "SELECT count(*) FROM knownGene" DB.from(:knownGene).count # => 82960 # instead of "SELECT name, chrom, txStart FROM knownGene LIMIT 1" DB.from(:knownGene).select(:name, :chrom, :txStart).first # => {:name=>"uc001aaa.3", :chrom=>"chr1", :txStart=>11873} # instead of "SELECT name FROM knownGene WHERE chrom == ’chrM’" DB.from(:knownGene).where(:chrom => "chrM").all # => [{:name=>"uc004coq.4"}, {:name=>"uc022bqo.2"}, {:name=>"uc004cor.1"}, {:name=>"uc004cos.5"}, # {:name=>"uc022bqp.1"}, {:name=>"uc022bqq.1"}, {:name=>"uc022bqr.1"}, {:name=>"uc031tga.1"}, # {:name=>"uc022bqs.1"}, {:name=>"uc011mfi.2"}, {:name=>"uc022bqt.1"}, {:name=>"uc022bqu.2"}, # {:name=>"uc004cov.5"}, {:name=>"uc031tgb.1"}, {:name=>"uc004cow.2"}, {:name=>"uc004cox.4"}, # {:name=>"uc022bqv.1"}, {:name=>"uc022bqw.1"}, {:name=>"uc022bqx.1"}, {:name=>"uc004coz.1"}]
- 8. Databases: Slide 8 of 24 don’t want to CREATE? you still might want to SELECT Question: How to map a SNP to a gene around +/- 60KB ? I am looking at a bunch of SNPs. Some of them are part of genes, but other are not. I am interested to look up +60KB or -60KB of those SNPs to get details about some nearby genes. Please share your experience in dealing with such a situation or thoughts on any methods that can do this. Thanks in advance. http://www.biostars.org/p/413/
- 9. Databases: Slide 9 of 24 example SELECT mysql --user=genome --host=genome-mysql.cse.ucsc.edu -A -D hg19 -e ’ select K.proteinID, K.name, S.name, S.avHet, S.chrom, S.chromStart, K.txStart, K.txEnd from snp130 as S left join knownGene as K on (S.chrom = K.chrom and not(K.txEnd + 60000 < S.chromStart or S.chromEnd + 60000 < K.txStart)) where S.name in ("rs25","rs100","rs75","rs9876","rs101") ’
- 10. Databases: Slide 10 of 24 example SELECT result
- 11. Databases: Slide 11 of 24 let’s talk about noSQL http://www.infoivy.com/2013/07/nosql-database-comparison-chart-only.html
- 12. Databases: Slide 12 of 24 (potentially) a good fit for biological data
- 13. Databases: Slide 13 of 24 many data sources are “key-value ready” (or close enough) http://togows.dbcls.jp/entry/pathway/hsa00030/genes.json [ { "2821": "GPI; glucose-6-phosphate isomerase [KO:K01810] [EC:5.3.1.9]", "2539": "G6PD; glucose-6-phosphate dehydrogenase [KO:K00036] [EC:1.1.1.49]", "25796": "PGLS; 6-phosphogluconolactonase [KO:K01057] [EC:3.1.1.31]", ... "5213": "PFKM; phosphofructokinase, muscle [KO:K00850] [EC:2.7.1.11]", "5214": "PFKP; phosphofructokinase, platelet [KO:K00850] [EC:2.7.1.11]", "5211": "PFKL; phosphofructokinase, liver [KO:K00850] [EC:2.7.1.11]" } ]
- 14. Databases: Slide 14 of 24 schema-free: save first, worry later (= agile) #!/usr/bin/ruby require "mongo" require "json/pure" require "open-uri" db = Mongo::Connection.new.db(’kegg’) col = db.collection(’genes’) j = JSON.parse(open("http://togows.dbcls.jp/entry/pathway/hsa00030/genes.json").read) j.each do |g| gene = Hash.new g.each_pair do |key, val| gene[:_id] = key gene[:desc] = val col.save(gene) end end Ruby code to save JSON from the TogoWS REST service
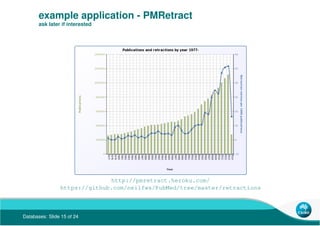
- 15. Databases: Slide 15 of 24 example application - PMRetract ask later if interested http://pmretract.heroku.com/ https://github.com/neilfws/PubMed/tree/master/retractions
- 16. Databases: Slide 16 of 24 when rows + columns != database - sometimes a database is overkill
- 17. Databases: Slide 17 of 24 example 1 - R/IRanges
- 18. Databases: Slide 18 of 24 example 2 - bedtools http://bedtools.readthedocs.org/en/latest/
- 19. Databases: Slide 19 of 24 example 3 - unix join (and the shell in general)
- 20. Databases: Slide 20 of 24 when are databases good? - when data are updated frequently - when multiple users do the updating - when queries are complex or ever-changing - as backends to web applications
- 21. Databases: Slide 21 of 24 when are databases not/less good? - for basic “set operations” - for sequence data [1] (?) [1] no time to discuss BioSQL, GBrowse/Bio::DB::GFF, BioDAS etc.
- 22. Databases: Slide 22 of 24 so how did I answer that email? options(java.parameters = "-Xmx4g") library(XLConnect) wb <- loadWorkbook("˜/Downloads/NGS Target list Tumour for Neil.xlsx") s1 <- readWorksheet(wb, sheet = 1, startCol = 1, endCol = 1, header = F) s2 <- readWorksheet(wb, sheet = 2, startCol = 1, endCol = 32, header = T) s4 <- readWorksheet(wb, sheet = 4, startCol = 1, endCol = 3, header = T) # then use gsub, match, %in% etc. to clean and join the data # ... Read spreadsheet into R using the XLConnect package, then “munge”






![Databases: Slide 7 of 24
my one tip for today: use ORM
= object relational mapping
#!/usr/bin/ruby
require ’sequel’
# connect to UCSC Genomes MySQL server
DB = Sequel.connect(:adapter => "mysql", :host => "genome-mysql.cse.ucsc.edu",
:user => "genome", :database => "hg19")
# instead of "SELECT count(*) FROM knownGene"
DB.from(:knownGene).count
# => 82960
# instead of "SELECT name, chrom, txStart FROM knownGene LIMIT 1"
DB.from(:knownGene).select(:name, :chrom, :txStart).first
# => {:name=>"uc001aaa.3", :chrom=>"chr1", :txStart=>11873}
# instead of "SELECT name FROM knownGene WHERE chrom == ’chrM’"
DB.from(:knownGene).where(:chrom => "chrM").all
# => [{:name=>"uc004coq.4"}, {:name=>"uc022bqo.2"}, {:name=>"uc004cor.1"}, {:name=>"uc004cos.5"},
# {:name=>"uc022bqp.1"}, {:name=>"uc022bqq.1"}, {:name=>"uc022bqr.1"}, {:name=>"uc031tga.1"},
# {:name=>"uc022bqs.1"}, {:name=>"uc011mfi.2"}, {:name=>"uc022bqt.1"}, {:name=>"uc022bqu.2"},
# {:name=>"uc004cov.5"}, {:name=>"uc031tgb.1"}, {:name=>"uc004cow.2"}, {:name=>"uc004cox.4"},
# {:name=>"uc022bqv.1"}, {:name=>"uc022bqw.1"}, {:name=>"uc022bqx.1"}, {:name=>"uc004coz.1"}]](https://image.slidesharecdn.com/foam2014db-140328070023-phpapp02/85/SQL-noSQL-or-no-database-at-all-Are-databases-still-a-core-skill-7-320.jpg)




![Databases: Slide 13 of 24
many data sources are “key-value ready”
(or close enough)
http://togows.dbcls.jp/entry/pathway/hsa00030/genes.json
[
{
"2821": "GPI; glucose-6-phosphate isomerase [KO:K01810] [EC:5.3.1.9]",
"2539": "G6PD; glucose-6-phosphate dehydrogenase [KO:K00036] [EC:1.1.1.49]",
"25796": "PGLS; 6-phosphogluconolactonase [KO:K01057] [EC:3.1.1.31]",
...
"5213": "PFKM; phosphofructokinase, muscle [KO:K00850] [EC:2.7.1.11]",
"5214": "PFKP; phosphofructokinase, platelet [KO:K00850] [EC:2.7.1.11]",
"5211": "PFKL; phosphofructokinase, liver [KO:K00850] [EC:2.7.1.11]"
}
]](https://image.slidesharecdn.com/foam2014db-140328070023-phpapp02/85/SQL-noSQL-or-no-database-at-all-Are-databases-still-a-core-skill-13-320.jpg)
![Databases: Slide 14 of 24
schema-free: save first, worry later
(= agile)
#!/usr/bin/ruby
require "mongo"
require "json/pure"
require "open-uri"
db = Mongo::Connection.new.db(’kegg’)
col = db.collection(’genes’)
j = JSON.parse(open("http://togows.dbcls.jp/entry/pathway/hsa00030/genes.json").read)
j.each do |g|
gene = Hash.new
g.each_pair do |key, val|
gene[:_id] = key
gene[:desc] = val
col.save(gene)
end
end
Ruby code to save JSON from the TogoWS REST service](https://image.slidesharecdn.com/foam2014db-140328070023-phpapp02/85/SQL-noSQL-or-no-database-at-all-Are-databases-still-a-core-skill-14-320.jpg)





![Databases: Slide 21 of 24
when are databases not/less good?
- for basic “set operations”
- for sequence data [1]
(?)
[1] no time to discuss BioSQL, GBrowse/Bio::DB::GFF, BioDAS etc.](https://image.slidesharecdn.com/foam2014db-140328070023-phpapp02/85/SQL-noSQL-or-no-database-at-all-Are-databases-still-a-core-skill-21-320.jpg)
