Lecture 10
- 1. Lecture 10 Enzyme Kinetics
- 2. Rate constants and reaction order Rate constant (k) measures how rapidly a reaction occurs k1 k-1 A B + C Rate (v, velocity) = (rate constant) (concentration of reactants) v= k1 [A] 1st order reaction (rate dependent on concentration of 1 reactant) v= k-1[B][C] 2nd order reaction (rate dependent on concentration of 2 reactants) Zero order reaction (rate is independent of reactant concentration)
- 3. Sample questions The rate of a second order reaction depends on the concentration of _________. • (a) one substrate • (b) two substrates • (c) three substrates • (d) none of the above
- 4. E + S E S E + P k1 k-1 k2 k-2 E + S ES E + P
- 5. Initial Velocities [S] = 1 mM Hold [E] constant [E]<<<<<[S] d[P]/dT = Vo1 mM [P] time
- 6. Initial Velocities d[P]/dT = Vo [S] = 10 mM 10 mM [S] = 5 mM [S] = 1 mM d[P]/dT = Vo5 mM d[P]/dT = Vo1 mM [P] time
- 7. Plot Vo vs. [S] Vo 5 mM Vo 1 mM Vo 10 mM
- 8. Initial Velocity Assumption 1) Measurements made to measure initial velocity (vo). At vo very little product formed. Therefore, the rate at which E + P react to form ES is negligible and k-2 is 0. Therefore E + S E S k1 k-1 k2 E + S ES E + P k-2 E + P
- 9. Steady State Assumption Steady state Assumption = [ES] is constant. The rate of ES formation equals the rate of ES breakdown E + S E S E + P k1 k-1 k2 E + S ES E + P
- 10. Data from a single experiment performed with at a single [S]. (single point on Vo vs. [S] plot)
- 11. Rate of ES formation E + S E S k1 E + S ES Rate = k1 [E] [S]
- 12. Rate of ES breakdown E S E + P k2 ES E + P E S E + S k-1 ES E + S Rate = (k2 [ES]) + (k-1[ES]) Rate = [ES](k2 + k-1)
- 13. If the rate of ES formation equals the rate of ES breakdown 1) k1[E][S] = [ES](k-1+ k2) 2) (k-1+ k2) / k1 = [E][S] / [ES] 3) (k-1+ k2) / k1 = Km (Michaelis constant)
- 14. Not required to know Michaelis-Menton Derivation k1 k2 k-1 E + S ES E + P 1. The overall rate of product formation: v = k2 [ES] 2. Rate of formation of [ES]: vf = k1[E][S] 3. Rate of decomposition of [ES]: vd = k-1[ES] + k2 [ES] 4. Rate of ES formation = Rate of ES decomposition (steady state) 5. So: k1[E][S] = k-1[ES] + k2 [ES]
- 15. Michaelis-Menton Derivation 6. In solving for [ES], use the enzyme balance to eliminate [E]. ET = [E] + [ES] 7. k1 (ET - [ES])[S] = k-1[ES] + k2 [ES] k1 ET[S] - k1[ES][S] = k-1[ES] + k2 [ES] 8. Rearrange and combine [ES] terms: k1 ET[S] = (k-1 + k2 + k1 [S])[ES] k1 ET[S] 9. Solve for [ES] = ----------------------- (k-1 + k2 + k1 [S]) Not required to know
- 16. Michaelis-Menton Derivation ET[S] 10. Divide through by k1: [ES] = ----------------------- (k-1 + k2)/k1 + [S] 11. Defined Michaelis constant: KM = (k-1 + k2) / k1 12. Substitute KM into the equation in step 10. 13. Then substitute [ES] into v = k2 [ES] from step1 and replace Vmax with k2 ET to give: Vmax[S] vo = ----------- KM + [S] Not required to know
- 17. Vmax = velocity where all of the enzyme is bound to substrate (enzyme is saturated with S) Km = [S] at ½ Vmax (units moles/L=M) (1/2 of enzyme bound to S)
- 18. Understanding Vmax The theoretical maximal velocity • Vmax is a constant • Vmax is the theoretical maximal rate of the reaction - but it is NEVER achieved in reality • To reach Vmax would require that ALL enzyme molecules are tightly bound with substrate • Vmax is asymptotically approached as substrate is increased
- 19. What does Km mean? 1. Km = [S] at ½ Vmax 2. Km is a constant; Km is a combination of rate constants describing the formation and breakdown of the ES complex 3. Km is usually a little higher than the physiological [S]
- 20. What does Km mean? 4. Km represents the amount of substrate required to bind ½ of the available enzyme (binding constant of the enzyme for substrate) 5. Km can be used to evaluate the specificity of an enzyme for a substrate 6. Small Km means tight binding; high Km means weak binding Glucose Km = 8 X 10-6 Allose Km = 8 X 10-3 Mannose Km = 5 X 10-6 Hexose Kinase Glucose + ATP <-> Glucose-6-P + ADP
- 23. Sample questions • How does the Michaelis-Menten equation explain why the rate of an enzyme-catalyzed reaction reaches a maximum value at high substrate? • At high So, Km <<<< So (numerically), so the term Km + So in the M-M equation becomes equal to So. Vo = (Vmax So)/So, and So cancels. Therefore at high So then, Vo = Vmax.
- 24. The turnover number A measure of catalytic activity • kcat, the turnover number, is the number of substrate molecules converted to product per enzyme molecule per unit of time, when E is saturated with substrate. • If the Michaelis-Menten model fits, k2 = kcat = Vmax/Et
- 25. What does kcat mean? 1. kcat is the 1st order rate constant describing ES E+P 2. Also known as the turnover number because it describes the number of reactions that a molecule of enzyme can catalyze per second under optimal condition. 3. Most enzyme have kcat values between 102 and 103 s-1 4. For simple reactions k2 = kcat , for multistep reactions kcat = rate limiting step k1 k-1 kcat E + S ES E + P
- 27. The catalytic efficiency What does kcat/Km mean? • It measures how the enzyme performs when S is low • kcat/Km describes an enzymes preference for different substrates = specificity constant • The upper limit for kcat/Km is the diffusion limit - substrate diffuse into the active site, or product diffuse out • Catalytic perfection when kcat/Km = diffusion rate
- 28. kcat/KM kcat/KM is taken to be a measure of the efficiency of an enzyme. Rewriting kcat/KM in terms of the kinetic constants gives: kcat k1k2 ---- = ----------- KM k-1 + k2 So, where k2 is small, the denominator becomes k-1 and kcat/KM is small.
- 30. Short summary • Km substrate specificity; substrate binding • kcat, the turnover number • kcat/Km the catalytic efficiency
- 31. Sample questions Which of the following kinetic parameters best describes how well suited a specific compound functions as a substrate for a particular enzyme? • (a) Km • (b) Vmax • (c) kcat • (d) kcat/Km
- 32. Sample questions The rate-determining step of Michaelis Menten kinetics is • A.the complex formation step • B.the complex dissociation step to produce product • C.the product formation step • D.Both (a)and(c)
- 33. Limitations of Michaelis-Menten model 1. Some enzyme catalyzed reactions show more complex behavior E + S<->ES<->EZ<->EP<-> E + P Michaelis-Menten can look only at rate limiting step 2. Often more than one substrate E+S1<->ES1+S2<->ES1S2<->EP1P2<-> EP2+P1<-> E+P2 Must optimize one substrate then calculate kinetic parameters for the other 3. Assumes k-2 = 0 4. Assume steady state conditions
- 34. The dual nature of the Michaelis-Menten equation Combination of 0-order and 1st-order kinetics • When S is low, the equation for rate is 1st order in S • When S is high, the equation for rate is 0- order in S • The Michaelis-Menten equation describes a hyperbolic dependence of v on S
- 35. How do you get values for Vmax, Km and kcat? • Can determine Km and Vmax experimentally • Km can be determined without an absolutely pure enzyme • Kcat values can be determined if Vmax is known and the absolute concentration of enzyme is known (Vmax = kcat[Etotal]
- 36. Vo [S] B B B B B B B 0.25 0.2 0.15 0.1 0.05 0 0 1 2 3 4 5 6 7 8 9 10 V max [S] Vo 0.5 0.075 0.75 0.09 2 0.152 4 0.196 6 0.21 8 0.214 10 0.23 Km Km ~ 1.3 mM Vmax ~ 0.25
- 37. Lineweaver-Burke Plots (double reciprocal plots) •Plot 1/[S] vs 1/Vo •L-B equation for straight line •X-intercept = -1/Km •Y-intercept = 1/Vmax •Easier to extrapolate values w/ straight line vs hyperbolic curve
- 38. Sample questions • For an enzyme (5 μM) , the following initial velocities have been reported depending on the substrate concentration: • (a) Draw a Michaelis-Menten plot for this enzyme. • (b) Draw a Lineweaver-Burke plot for this enzyme. • (c) Determine Km and Vmax for this enzyme • (d) Indicate in both graphs (a & b) where Vmax and Km can be recognized. • (e) Calculate the turnover number and the catalytic efficiency for this enzyme. [Substrate], mM v0, mM/s 0.02 10.83 0.04 18.57 0.07 26.76 0.1 32.50 0.15 39.00 0.2 43.33 0.3 48.75 0.5 54.17 0.7 56.88 kcat = Vmax / [E]total catalytic efficiency: kcat/Km
- 39. Answer • (a) • (b) • (c) Km and Vmax can be determined from the intercepts in the Lineweaver- Burke plot: 1/Vmax = 0.015 s/mM Vmax = 66 mM/s -1/Km = -10 mM Km = 0.1 mM • (e) kcat = Vmax / [E]total = 65 mM/s ÷ 5 μM = 65 mM/s ÷ 0.005 mM = 13000/s catalytic efficiency: kcat/Km = 13000/s ÷ 0.1 mM = 13000/s ÷ 0.0001 M = 1.3×108 M/s
- 40. Enzyme Inhibition • Inhibitor – substance that binds to an enzyme and interferes with its activity • Can prevent formation of ES complex or prevent ES breakdown to E + P. Reversible versus Irreversible • Reversible inhibitors interact with an enzyme via noncovalent associations • Irreversible inhibitors interact with an enzyme via covalent associations
- 42. Reversible Inhibitors E + S <-> ES -> E + P E + I <-> EI Ki = [E][I]/[EI] • Competitive • Uncompetitive • Non-competitive
- 43. Classes of Reversible Inhibition Two real, one hypothetical • Competitive inhibition - inhibitor (I) competes with the substrate for the active site of the enzyme • Non-competitive inhibition - inhibitor (I) binds to an enzyme somewhere other than the active site. It can binds either ES or E. A non-competitive inhibitor reacts with the enzyme-substrate complex, and slows the rate of reaction to form the enzyme-product complex. • Uncompetitive inhibition - inhibitor (I) binds only to ES, not to E. This is a hypothetical case that has never been documented for a real enzyme, but which makes a useful contrast to competitive inhibition
- 47. • Competitive inhibitor: Vmax stays the same, but Km increases • Non-competitive inhibitor decreases the turnover number of the enzyme rather than preventing substrate binding- Vmax decreases but Km stays the same. This cannot be overcome with an increase in substrate concentration.
- 48. Enzyme Inhibition Noncovalent binding: Competitive (I binds only to E) Uncompetitive (I binds only to ES) Noncompetitive (I binds to E or ES) Covalent binding – irreversible Group Specific Substrate Analogs (bound to the active site and prevent further reactions)
- 49. Competitive Inhibitor (CI) •CI binds free enzyme •Competes with substrate for enzyme binding. •Raises Km without effecting Vmax •Can relieve inhibition with more S
- 50. Competitive Inhibitors look like substrate O HO C NH2 S NH2 O H2N O PABA Sulfanilamide
- 51. Non-competitive Inhibitor (NI) •NI can bind free E or ES complex •Lowers Vmax, but Km remains the same •NI’s don’t bind to S binding site therefore don’t effect Km •Alters conformation of enzyme to effect catalysis but not substrate binding
- 52. Uncompetitive Inhibitor (UI) •UI binds ES complex •Prevents ES from proceeding to E + P or back to E + S. •Lowers Km & Vmax, but ratio of Km/Vmax remains the same •Occurs with multisubstrate enzymes
- 53. Sample questions Which of the following binds to an enzyme at its active site? • A) irreversible inhibitor • B) reversible competitive inhibitor • C) reversible noncompetitive inhibitor • D) more than one correct response • E) no correct response An uncompetitive inhibitor binds to _____. • (a) E • (b) ES • (c) P • (d) a and b • (e) a and c
- 54. Sample questions A reversible inhibitor that can bind to either E alone or the ES complex is referred to as a _____. • (a) competitive inhibitor. • (b) non-competitive inhibitor. • (c) uncompetitive inhibitor. • (d) suicide inhibitor. • (e) irreversible inhibitor.
- 55. Sample questions A competitive inhibitor of an enzyme is usually • A.a highly reactive compound • B.a metal ion such as Hg2+ or Pb2+ • C.structurally similar to the substrate. • D.water insoluble The enzyme inhibition can occur by • A.reversible inhibitors • B.irreversible inhibitors • C.Both (a) and (b) • D.None of these
- 56. Sample questions In a Lineweaver-Burk Plot, competitive inhibitor shows which of the following effect? • A.It moves the entire curve to right • B.It moves the entire curve to left • C.It changes the x-intercept • D.It has no effect on the slope
- 57. Sample questions Non-competitive inhibitor of an enzyme catalyzed reaction • A.decreases Vmax • B.binds to ES • C.both (a) and (b) • D.can actually increase reaction velocity in rare cases
- 58. Sample questions A classical uncompetitive inhibitor is a compound that binds • A.reversibly to the enzyme substrate complex yielding an inactive ESI complex • B.irreversibly to the enzyme substrate complex yielding an inactive ESI complex • C.reversibly to the enzyme substrate complex yielding an active ESI complex • D.irreversibly to the enzyme substrate complex yielding an active ESI complex
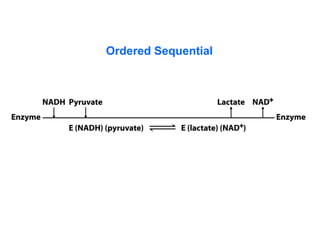
- 59. Kinetics of Multisubstrate Reactions E + A + B <-> E + P + Q • Sequential Reactions a) ordered b) random • Ping-pong Reactions
- 60. Sequential Reactions A B P Q E EA (EAB) (EPQ) EQ E A B B A Q P E EA EB (EAB)(EPQ) P Q EQ EP E Ordered Random
- 62. Ordered Random
- 63. Ping-Pong Reactions A P B Q E (EA)(FP) (F) (FB)(EQ) E •In Ping-Pong reactions first product released before second substrate binds •When E binds A, E changes to F •When F binds B, F changes back to E
- 65. Lineweaver-Burke Plot of Multisubstrate Reactions Increasing [B] Increasing [B] Sequential Ping-Pong Vmax doesn’t change Km changes Both Vmax & Km change 1/Vo 1/[S] 1/Vo 1/[S]


![Rate constants and reaction order
Rate constant (k) measures how rapidly a reaction occurs
k1
k-1
A B + C
Rate (v, velocity) = (rate constant) (concentration of reactants)
v= k1 [A]
1st order reaction (rate dependent on concentration of 1 reactant)
v= k-1[B][C]
2nd order reaction (rate dependent on concentration of 2 reactants)
Zero order reaction (rate is independent of reactant concentration)](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-2-320.jpg)


![Initial Velocities
[S] = 1 mM
Hold [E] constant
[E]<<<<<[S]
d[P]/dT = Vo1 mM
[P]
time](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-5-320.jpg)
![Initial Velocities
d[P]/dT = Vo [S] = 10 mM 10 mM
[S] = 5 mM
[S] = 1 mM
d[P]/dT = Vo5 mM
d[P]/dT = Vo1 mM
[P]
time](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-6-320.jpg)
![Plot Vo vs. [S]
Vo 5 mM
Vo 1 mM
Vo 10 mM](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-7-320.jpg)

![Steady State Assumption
Steady state Assumption = [ES] is constant. The rate of ES
formation equals the rate of ES breakdown
E + S E S E + P
k1
k-1
k2
E + S ES E + P](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-9-320.jpg)
![Data from a single experiment
performed with at a single [S].
(single point on Vo vs. [S] plot)](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-10-320.jpg)
![Rate of ES formation
E + S E S
k1
E + S ES
Rate = k1 [E] [S]](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-11-320.jpg)
![Rate of ES breakdown
E S E + P
k2
ES E + P
E S E + S
k-1
ES E + S
Rate = (k2 [ES]) + (k-1[ES])
Rate = [ES](k2 + k-1)](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-12-320.jpg)
![If the rate of ES formation equals the rate of ES
breakdown
1) k1[E][S] = [ES](k-1+ k2)
2) (k-1+ k2) / k1 = [E][S] / [ES]
3) (k-1+ k2) / k1 = Km (Michaelis constant)](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-13-320.jpg)
![Not required to know
Michaelis-Menton Derivation
k1 k2
k-1
E + S ES E + P
1. The overall rate of product formation: v = k2 [ES]
2. Rate of formation of [ES]: vf = k1[E][S]
3. Rate of decomposition of [ES]:
vd = k-1[ES] + k2 [ES]
4. Rate of ES formation = Rate of ES decomposition
(steady state)
5. So: k1[E][S] = k-1[ES] + k2 [ES]](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-14-320.jpg)
![Michaelis-Menton Derivation
6. In solving for [ES], use the enzyme balance to
eliminate [E]. ET = [E] + [ES]
7. k1 (ET - [ES])[S] = k-1[ES] + k2 [ES]
k1 ET[S] - k1[ES][S] = k-1[ES] + k2 [ES]
8. Rearrange and combine [ES] terms:
k1 ET[S] = (k-1 + k2 + k1 [S])[ES]
k1 ET[S]
9. Solve for [ES] = -----------------------
(k-1 + k2 + k1 [S])
Not required to know](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-15-320.jpg)
![Michaelis-Menton Derivation
ET[S]
10. Divide through by k1: [ES] = -----------------------
(k-1 + k2)/k1 + [S]
11. Defined Michaelis constant: KM = (k-1 + k2) / k1
12. Substitute KM into the equation in step 10.
13. Then substitute [ES] into v = k2 [ES] from step1
and replace Vmax with k2 ET to give:
Vmax[S]
vo = -----------
KM + [S]
Not required to know](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-16-320.jpg)
![Vmax = velocity where all of the
enzyme is bound to substrate
(enzyme is saturated with S)
Km = [S] at ½ Vmax
(units moles/L=M)
(1/2 of enzyme bound to S)](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-17-320.jpg)

![What does Km mean?
1. Km = [S] at ½ Vmax
2. Km is a constant; Km is a combination of rate
constants describing the formation and breakdown
of the ES complex
3. Km is usually a little higher than the physiological [S]](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-19-320.jpg)















![How do you get values for Vmax, Km and kcat?
• Can determine Km and Vmax experimentally
• Km can be determined without an absolutely pure
enzyme
• Kcat values can be determined if Vmax is known and
the absolute concentration of enzyme is known (Vmax
= kcat[Etotal]](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-35-320.jpg)
![Vo [S]
B
B
B
B
B B
B
0.25
0.2
0.15
0.1
0.05
0
0 1 2 3 4 5 6 7 8 9 10
V max
[S] Vo
0.5 0.075
0.75 0.09
2 0.152
4 0.196
6 0.21
8 0.214
10 0.23
Km
Km ~ 1.3 mM
Vmax ~ 0.25](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-36-320.jpg)
![Lineweaver-Burke Plots
(double reciprocal plots)
•Plot 1/[S] vs 1/Vo
•L-B equation for straight
line
•X-intercept = -1/Km
•Y-intercept = 1/Vmax
•Easier to extrapolate
values w/ straight line vs
hyperbolic curve](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-37-320.jpg)
![Sample questions
• For an enzyme (5 μM) , the following
initial velocities have been reported
depending on the substrate
concentration:
• (a) Draw a Michaelis-Menten plot for
this enzyme.
• (b) Draw a Lineweaver-Burke plot for
this enzyme.
• (c) Determine Km and Vmax for this
enzyme
• (d) Indicate in both graphs (a & b)
where Vmax and Km can be
recognized.
• (e) Calculate the turnover number and
the catalytic efficiency for this enzyme.
[Substrate],
mM
v0, mM/s
0.02 10.83
0.04 18.57
0.07 26.76
0.1 32.50
0.15 39.00
0.2 43.33
0.3 48.75
0.5 54.17
0.7 56.88
kcat = Vmax / [E]total catalytic efficiency: kcat/Km](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-38-320.jpg)
![Answer
• (a)
• (b)
• (c) Km and Vmax can be determined from the intercepts in the Lineweaver-
Burke plot:
1/Vmax = 0.015 s/mM Vmax = 66 mM/s
-1/Km = -10 mM Km = 0.1 mM
• (e) kcat = Vmax / [E]total = 65 mM/s ÷ 5 μM
= 65 mM/s ÷ 0.005 mM = 13000/s
catalytic efficiency: kcat/Km = 13000/s ÷ 0.1 mM
= 13000/s ÷ 0.0001 M
= 1.3×108 M/s](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-39-320.jpg)


![Reversible Inhibitors
E + S <-> ES -> E + P
E + I <-> EI
Ki = [E][I]/[EI]
• Competitive
• Uncompetitive
• Non-competitive](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-42-320.jpg)





















![Lineweaver-Burke Plot of
Multisubstrate Reactions
Increasing
[B]
Increasing
[B]
Sequential Ping-Pong
Vmax doesn’t change
Km changes
Both Vmax & Km change
1/Vo
1/[S]
1/Vo
1/[S]](https://image.slidesharecdn.com/4j0ncajgqqml2lkdvrdm-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate02/85/Lecture-10-65-320.jpg)