Suppressor mutation
- 1. Suppressor Mutation Name- DeepikaRana RollNo.-1601 Deptt.- Microbiology(2nd ) M.D. University, Rohtak
- 2. •A suppressor mutation is a mutation that counters the phenotypic effect of a previous mutation. •It may either alleviates or reverts the phenotypic effects of an already existing mutation. •Genetic suppression therefore restores the phenotype seen prior to the original background mutation.
- 3. For example, say a mutation causes the codon 'AGA' (which codes for the amino acid Arginine) to change to 'AGC' (which codes for the amino acid Serine, a very different amino acid to Arginine). A suppressor mutation may be one that changes the mutated codon to 'CGC' (which also codes for Arginine), or to 'AAG' (which codes for Lysine, a similar amino acid to Arginine), thereby reversing the effect the first mutation may have had on the function of the protein.
- 7. (a) A typical receptor-mediated signal transduction pathway is shown, involving a ligand, a cell surface receptor, a G protein, a kinase cascade and a transcription factor that activates or represses gene expression in response to the signal. (b) A null mutation in the gene encoding kinase 1 blocks the pathway, resulting in a mutant phenotype (i). The null mutation can be suppressed by a mutation in a gene encoding a component (denoted by an asterisk), such as kinase 3 (ii), which activates the downstream portion of the pathway independent of kinase 1 activity. Mutations in upstream components, such as the G protein (iii), restore signaling through only part of the pathway, owing to the kinase 1 defect. (c) When the pathway is inactivated by a partial loss-of-function mutation in kinase 1 (i), mutations in either downstream (ii) or upstream (iii) components can activate the pathway enough to suppress the mutant phenotype. Pathways can, therefore, only be ordered reliably when the starting mutation is a null allele.
- 8. Intragenic suppression •Intragenic suppression, where a phenotype caused by a primary mutation is ameliorated by a second mutation in the same gene. • The suppressing mutation might be a true revertant, restoring the original DNA sequence; it might be an alteration of the same codon, resulting in a less detrimental amino acid at that position; or it might affect a different codon, causing an amino acid change at another position that now restores the function of that protein closer to wild-type activity. •Intragenic suppressors must be very tightly linked to the original mutation, whereas extragenic suppressors are unlikely to be tightly linked. •Although intragenic suppressors provide valuable information about the structure–function relationships within a protein, they do not identify any new proteins that are functionally related to the original mutant, and therefore are generally not the goal when undertaking a suppressor hunt.
- 9. Intergenic Suppression • Also known as extragenic suppression relieves the effects of a mutation in one gene by a mutation somewhere else within the genome. •The second mutation is not on the same gene as the original mutation. Intergenic suppression is useful for identifying and studying interactions between molecules, such as proteins. • For example, a mutation which disrupts the complementary interaction between protein molecules may be compensated for by a second mutation elsewhere in the genome that restores or provides a suitable alternative interaction between those molecules. •Several proteins of biochemical, signal transduction, and gene expression pathways have been identified using this approach. •Examples of such pathways include receptor- ligand interactions as well as the interaction of components involved in DNA replication, transcription, and translation.
- 10. Suppression of frameshift mutations. (a) A deletion in the nucleotide coding sequence can result in an incomplete, inactive polypeptide chain. (b) The effect of the deletion, shown in figure a, can be overcome by a second mutation, an insertion in the coding sequence. This insertion results in the production of a complete polypeptide chain having two amino acid replacements. These may revert by the restoration of a deleted base for example, but can also be suppressed by the addition or deletion of further bases (not necessarily at the same place as the original mutation) so that the total number of bases added or lost is a multiple of three In this way the original reading frame is restored leaving a limited number of altered codons. Whether this altered product has sufficient biological function to result in observable suppression of the original mutation will of course depend on the size and nature of this altered sequence and its effect on the function of the protein.
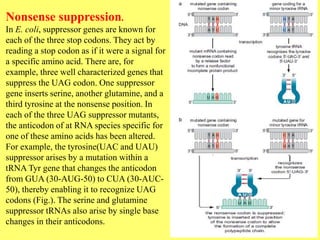
- 11. Nonsense suppression. In E. coli, suppressor genes are known for each of the three stop codons. They act by reading a stop codon as if it were a signal for a specific amino acid. There are, for example, three well characterized genes that suppress the UAG codon. One suppressor gene inserts serine, another glutamine, and a third tyrosine at the nonsense position. In each of the three UAG suppressor mutants, the anticodon of at RNA species specific for one of these amino acids has been altered. For example, the tyrosine(UAC and UAU) suppressor arises by a mutation within a tRNA Tyr gene that changes the anticodon from GUA (30-AUG-50) to CUA (30-AUC- 50), thereby enabling it to recognize UAG codons (Fig.). The serine and glutamine suppressor tRNAs also arise by single base changes in their anticodons.
- 12. Suppression of a nonsense mutation. A base substitution changes CAG to the stop codon UAG, causing pre-mature termination of translation. This can be suppressed by a separate mutation in a tRNA gene, giving rise to a tRNA that can recognize the UAG codon. The original mutation changes a glutamine codon (CAG) to a stop codon (UAG). Suppression of this mutation can occur by alteration of the glutamine tRNA gene so that its anticodon now pairs with the amber codon. Glutamine will therefore be inserted into the growing peptide chain and the final product will be identical with the wild-type protein. Since there is more than one glutamine tRNA gene, the cell does not lose the ability to recognize genuine glutamine codons. The importance of this type of suppression is that the tRNA mutation is able to suppress any corresponding mutation, not just the original one it was selected for.
- 13. •Suppression of nonsense mutations is a simple approach to study the effect of different amino acid substitutions at a specific position in a protein. Promega sells a nice kit of nonsense suppressors for this type of analysis. •Nonsense suppressors can also be used to incorporate unnatural amino acids into proteins. For example, alpha-hydroxy acids with a wide variety of substituent can be synthesized and attached to a nonsense suppressor tRNA, allowing substitution of the unnatural amino acid for any chosen amino acid residue in the protein. •The unnatural amino acids can provide a probe for the role of that position in the structure and function of the protein, etc.
- 14. Genetic tests for protein interactions. Suppressor mutations. Two proteins, A and B, normally interact. A mutation in A prevents the interaction, causing a loss of function phenotype, but this can be suppressed by a complementary mutation in B which restores the interaction. •To show potential interactions between proteins by screening for suppressor mutations, i.e. mutations in one gene that partially or fully compensate for a mutation in another. The primary mutation causes a change in protein structure that prevents the interaction, but the suppressor mutation introduces a complementary change in the second protein that restores it. widely employed in amenable organisms like Drosophila and yeast. •Advantage- provide a short cut to functionally significant interactions, sifting through the proteome for those interactions that have a recognizable effect on the overall phenotype
- 15. •The analysis of any biological process by classical genetic methods ultimately requires multiple types of mutant selections to identify all the genes involved in that process. •One strategy commonly used to identify functionally related genes is to begin with a strain that already contains a mutation affecting the pathway of interest, selecting for mutations that modify its phenotype. • Modifiers that result in a more severe phenotype are termed enhancers, while mutations that restore a more wild-type phenotype despite the continued presence of the original mutation are termed suppressors.
- 16. •They also provide evidence between functionally interacting molecules and intersecting biological pathways. •Suppressor analysis is a commonly used strategy to identify functional relationships between genes that might not have been revealed through other genetic or biochemical means. •Suppressors have proven extremely valuable for determining the relationship between two gene products in vivo, even in the absence of cloning or sequence information
- 17. There are two main reasons for the increased use of suppressors. •First, many genes are resistant to identification by more direct genetic selections. A pre-existing mutation often serves to sensitize that pathway, allowing the identification of additional components through suppressor selections. •Second, and perhaps more importantly, suppression of a pre-existing phenotype establishes a genetic relationship between the two genes that might not have been established by other methods.
- 18. •Molecular Genetics of Bacteria 4th Edition (2004) Jeremy W. DaleSimon F. Park University of Surrey, UK •Prescott Harley, and Klein’s Microbiology seventh edition (2008) Joanne M.Willey Hofstra University Linda M. Sherwood Montana State University Christopher J.Woolverton Kent State University •Principles of Gene Manipulation and Genomics SEVENTH EDITION (2006) S.B. Primrose and R.M. Twyman •Reversion and Suppression - SDSU College of Sciences www.sci.sdsu.edu/~smaloy/MicrobialGenetics/topics/rev-sup/ •Suppressor mutation - Wikipedia, the free encyclopedia https://en.wikipedia.org/wiki/Suppressor_mutation •Mutation www.cod.edu/people/faculty/fancher/genetics/Mutation.htm •Suppression mechanisms themes from variations(Review Article) Gregory Prelich Department of Molecular Genetics, Albert Einstein College of Medicine, 1300 Morris Park Avenue,Bronx, New York NY 10461, USA.