Mixed Effects Models - Data Processing
- 1. Week 2.2: Data Processing in R ! Filtering ! Basic Filtering ! Advanced Filtering ! Mutate ! Basic Variable Creation and Editing ! if_else() ! Variable Types ! Other Functions & Packages
- 2. Filtering Data ! We didn’t see a big difference between conditions ! But, some RTs look like outliers—we may want to exclude them
- 3. Filtering Data ! Often, we want to examine or use just part of a dataframe ! filter() lets us retain only certain observations ! experiment %>% filter(RT < 2000) %>% group_by(Condition) %>% summarize(M=mean(RT)) Inclusion criterion: We want to keep RTs less than 2000 ms As we saw last time, this gets the mean RT for each condition
- 4. Filtering Data ! Often, we want to examine or use just part of a dataframe ! filter() lets us retain only certain observations ! experiment %>% filter(RT < 2000) %>% group_by(Condition) %>% summarize(M=mean(RT)) Inclusion criterion: We want to keep RTs less than 2000 ms
- 5. Filtering Data ! This only temporarily filtered the data ! If we want to
- 6. Filtering Data ! This only temporarily filtered the data ! If we want to run a lot of analyses with this filter, we may want to save the filtered data as a new dataframe ! experiment %>% filter(RT < 2000) -> experiment.filtered -> is the assignment operator. It stores results or data in memory. Name of the new dataframe (can be whatever you want)
- 7. Filtering Data ! This only temporarily filtered the data ! If we want to run a lot of analyses with this filter, we may want to save the filtered data as a new dataframe
- 8. Writing Data ! Note that this is just creating a new dataframe in R ! If you want to save to a folder on your computer, use write.csv(): ! write.csv(experiment.filtered, file='experiment_filtered.csv')
- 9. Filtering Data ! Why not just delete the bad RTs from the spreadsheet?
- 10. Filtering Data ! Why not just delete the bad RTs from the spreadsheet? ! Easy to make a mistake / miss some of them ! Faster to have the computer do it ! We’d lose the original data ! No documentation of how we subsetted the data
- 11. Week 2.2: Data Processing in R ! Filtering ! Basic Filtering ! Advanced Filtering ! Mutate ! Basic Variable Creation and Editing ! if_else() ! Variable Types ! Other Functions & Packages
- 12. Filtering Data: AND and OR ! What if we wanted only RTs between 200 and 2000 ms? - experiment %>% filter(RT >= 200 & RT <= 2000) ! | means OR: - experiment %>% filter(RT < 200 | RT > 2000) -> experiment.outliers - Logical OR (“either or both”)
- 13. Filtering Data: == and != ! Get a match / equals: - experiment %>% filter(TrialsRemaining == 0) ! Words/categorical variables need quotes: - experiment %>% filter(Condition=='Implausible') ! != means “not equal to”: - experiment %>% filter(Subject != 'S23’) - Drops Subject “S23” Note DOUBLE equals sign
- 14. Filtering Data: %in% ! Sometimes our inclusion criteria aren't so mathematical ! Suppose I just want the “Ducks” and “Panther” items ! We can check against any arbitrary list: - experiment %>% filter(ItemName %in% c('Ducks', 'Panther')) ! Or, keep just things that aren't in a list: - experiment %>% filter(Subject %in% c('S10', 'S23') == FALSE)
- 15. Logical Operators Review ! Summary - > Greater than - >= Greater than or equal to - < Less than - <= Less than or equal to - & AND - | OR - == Equal to - != Not equal to - %in% Is this included in a list?
- 16. Week 2.2: Data Processing in R ! Filtering ! Basic Filtering ! Advanced Filtering ! Mutate ! Basic Variable Creation and Editing ! if_else() ! Variable Types ! Other Functions & Packages
- 17. Mutate ! The last tidyverse function we’ll look at is mutate() ! Add new variables ! Transform variables ! Recode or rescore variables
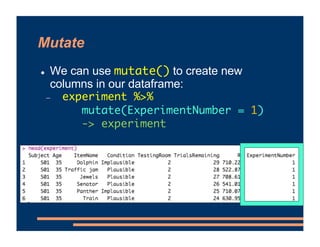
- 18. Mutate ! We can use mutate() to create new columns in our dataframe: - experiment %>% mutate(ExperimentNumber = 1) -> experiment We are creating a column named ExperimentNumber, and assigning the value 1 for every observation Then, we need to store the updated data back into our experiment dataframe
- 19. Mutate ! We can use mutate() to create new columns in our dataframe: - experiment %>% mutate(ExperimentNumber = 1) -> experiment
- 20. Mutate ! A more interesting example is where the assigned value is based on a formula ! experiment %>% mutate(RTinSeconds = RT/1000) -> experiment ! For each row, finds the RT in seconds for that specific trial and saves that into RTinSeconds - Similar to an Excel formula • If we wanted to alter the original RT column, we could instead do: mutate(RT = RT/1000)
- 21. Mutate ! We can even use other functions in calculating new columns ! experiment %>% mutate(logRT = log(RT)) -> experiment ! Applies the logarithmic transformation to each RT and saves that as logRT
- 22. Week 2.2: Data Processing in R ! Filtering ! Basic Filtering ! Advanced Filtering ! Mutate ! Basic Variable Creation and Editing ! if_else() ! Variable Types ! Other Functions & Packages
- 23. if_else() IF YOU WANT DESSERT, EAT YOUR PEAS … OR ELSE!
- 24. if_else() ! if_else(): A function that uses a test to decide which of two values to assign: ! experiment %>% mutate( Half= if_else( TrialsRemaining >= 15, 1, 2) ) -> experiment Function name If 15 or more trials remain… “Half” is 1 If NOT, “Half” is 2 A new column called “Half”--what value are we going to assign ?
- 25. Which do you like better? - experiment %>% mutate( Half=if_else(TrialsRemaining >= 15, 1, 2)) -> experiment ! vs: - TrialsPerSubject <- 30 - experiment %>% mutate( Half=if_else(TrialsRemaining >= TrialsPerSubject / 2, 1, 2)) -> experiment
- 26. Which do you like better? - experiment %>% mutate( Half=if_else(TrialsRemaining >= 15, 1, 2)) -> experiment ! vs: - TrialsPerSubject <- 30 - experiment %>% mutate( Half=if_else(TrialsRemaining >= TrialsPerSubject / 2, 1, 2)) -> experiment - Explains where the 15 comes from—helpful if we come back to this script later - We can also refer to CriticalTrialsPerSubject variable later in the script & this ensure it’s consistent - Easy to update if we change the number of trials
- 27. if_else() ! Instead of comparing to specific numbers (like 15), we can use other columns or a formula: ! experiment %>% mutate( RT.Fenced = if_else(RT < 200, 200, RT)) -> experiment ! What is this doing?
- 28. if_else() ! Instead of comparing to specific numbers (like 15), we can use other columns or a formula: ! experiment %>% mutate( RT.Fenced = if_else(RT < 200, 200, RT)) -> experiment ! Creates an RT.Fenced column where: ! Where RTs are less than 200 ms, replace them with 200 ! Otherwise, use the original RT value ! i.e., replace all RTs less than 200 ms with the value 200
- 29. if_else() ! Instead of comparing to specific numbers (like 15), we can use other columns or a formula: ! experiment %>% mutate( RT.Fenced = if_else(RT < 200, 200, RT)) -> experiment ! For even more complex rescoring, use case_when()
- 30. Week 2.2: Data Processing in R ! Filtering ! Basic Filtering ! Advanced Filtering ! Mutate ! Basic Variable Creation and Editing ! if_else() ! Variable Types ! Other Functions & Packages
- 31. Types ! R treats continuous & categorical variables differently: ! These are different data types: - Numeric - Character: Freely entered text (e.g., open response question) - Factor: Variable w/ fixed set of categories (e.g., treatment vs. placebo)
- 32. Types ! R’s current heuristic when reading in data: - No letters, purely numbers → numeric - Letters anywhere in the column → character
- 33. Types: as.factor() ! For variables with a fixed set of categories, we may want to convert to factor ! experiment %>% mutate(Condition=as.factor(Condition)) -> experiment
- 34. Types: as.numeric() ! Age was read as a character variable because some people “Declined to report” ! But, we may want to treat it as numeric despite this
- 35. Types: as.numeric() ! Age was read as a character variable because some people “Declined to report” - experiment %>% mutate(AgeNumeric=as.numeric(Age)) -> experiment • We now get quantitative information on Age • Values that couldn’t be turned into numbers are listed as NA • NA means missing data--we’ll discuss that more later in the term
- 36. Week 2.2: Data Processing in R ! Filtering ! Basic Filtering ! Advanced Filtering ! Mutate ! Basic Variable Creation and Editing ! if_else() ! Variable Types ! Other Functions & Packages
- 37. Other Functions ! Some built-in analyses: ! aov() ANOVA ! lm() Linear regression ! glm() Generalized linear models (e.g., logistic) ! cor.test() Correlation ! t.test() t-test
- 38. Other Packages ! Some other relevant packages: ! lavaan Latent variable analysis and structural equation modeling ! psych Psychometrics (scale construction, etc.) ! party Random forests ! stringr Working with character variables ! lme4: Package for linear mixed-effects models ! Get this one for next week
- 39. Getting Help ! Get help on a specific known function: - ?t.test - Lists all arguments ! Try to find a function on a particular topic: - ??logarithm